Thursday, 14 February 2019 04:10
Written by Caterina
A manuscript describing the OMEGA application was recently published on BiorXiv.org. For more details see: bioRxiv 251850; DOI: https://doi.org/10.1101/251850
Summary:
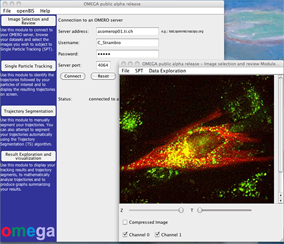
Open Microscopy Environment inteGrated Analysis (OMEGA) is a cross-platform data management, analysis, and visualization system, for particle tracking data, with particular emphasis on results from viral and vesicular trafficking experiments. OMEGA provides intuitive graphical interfaces to implement integrated particle tracking and motion analysis workflows while providing easy to use facilities to automatically keep track of error propagation, harvest data provenance and ensure the persistence of analysis results and metadata. Specifically, OMEGA: 1) imports image data and metadata from data management tools such as the Open Microscopy Environment Remote Objects (OMERO; Allan et al., 2012); 2) tracks intracellular particles movement; 3) facilitates parameter optimization and trajectory results inspection and validation; 4) performs downstream trajectory analysis and motion type classification; 5) estimates the uncertainty propagating through the motion analysis pipeline; and, 6) facilitates storage and dissemination of analysis results, and analysis definition metadata, on the basis of our newly proposed FAIRsharing.org complainant Minimum Information About Particle Tracking Experiments (MIAPTE; (Rigano and Strambio-De-Castillia, 2016; 2017) guidelines in combination with the OME-XML data model (Goldberg et al., 2005). In so doing, OMEGA maintains a persistent link between raw image data, intermediate analysis steps, the overall analysis output, and all necessary metadata to repeat the analysis process and reproduce its results.