Difference between revisions of "Software"
| (11 intermediate revisions by 2 users not shown) | |||
| Line 8: | Line 8: | ||
membrane of a cell). | membrane of a cell). | ||
| − | The following narratives describe some of the BIG software, including images and references. | + | The following narratives describe some of the BIG software, including images and references. The first 6 include a link to a separate web page which provides further information. |
This software was written, primarily, by BIG members | This software was written, primarily, by BIG members | ||
| Line 65: | Line 65: | ||
the latter of which can now restore images in seconds on a single workstation, | the latter of which can now restore images in seconds on a single workstation, | ||
make EPR for the first time amenable for High-Througput, high-speed and intelligent acquisition modalitis. | make EPR for the first time amenable for High-Througput, high-speed and intelligent acquisition modalitis. | ||
| + | |||
| + | <gallery heights=220px mode=packed-hover> | ||
| + | File:WF-demo-annotated.png|link=http://big.umassmed.edu/wiki/images/1/1e/WF-demo-annotated.png | ||
| + | File:SL-demo-annotated.png|link=http://big.umassmed.edu/wiki/images/8/81/SL-demo-annotated.png | ||
| + | File:SLeEPR-demo-annotated.gif|link=http://big.umassmed.edu/wiki/images/f/f3/SLeEPR-demo-annotated.gif | ||
| + | </gallery> | ||
References: | References: | ||
| Line 127: | Line 133: | ||
---- | ---- | ||
| + | |||
= I2I - The BIG Image File Format = | = I2I - The BIG Image File Format = | ||
The I2I image file format dates back to the mid-1980s, built on a format first created by Dr. James Coggins. | The I2I image file format dates back to the mid-1980s, built on a format first created by Dr. James Coggins. | ||
Latest revision as of 19:37, 29 September 2016
Biomedical Imaging Group Software Overview
Over the last three decades, the BIG has written and validated a large collection of software tools (hundreds of programs) for image acquisition, processing, analysis and visualization, image restoration, computer vision, and the modeling and simulation of biological systems. The bulk of this software is written in C, as well as in Fortran, C++, Perl, Java and Matlab. These programs range from the simple (e.g., add/subtract/multiply/divide images; perform global thresholding, etc.) to the complex (e.g., deformable models to identify the plasma membrane of a cell).
The following narratives describe some of the BIG software, including images and references. The first 6 include a link to a separate web page which provides further information.
This software was written, primarily, by BIG members
Lawrence Lifshitz (this includes an overview of Dr. Lifshitz's research areas and a full bibliography)
Shrinkwrap - a Deformable Surfaces program
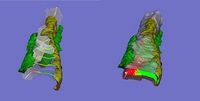
We have developed and implemented a deformable surface model called "shrinkwrap" which has been used numerous times over the years to locate the plasma membrane of fluorescently labeled cells. The model is composed of vertices spaced along a model surface, which is initially placed within a 3D image, near its correct location. Each vertex is then allowed move in response to image intensity gradients, attempting to move to locations in the image with a higher intensity while imposing a cost based upon how much curvature its position would impose onto the surface. Our implementation calculates curvature measures (i.e., first and second derivatives of the surface) and intensity measures locally and quickly. This results in a fast algorithm, which is also easy to modify to handle additional local constraints.
Other programs related to finding the surface of a smooth muscle cell and performing related analysis:
- Corecell.pl : Takes a 3D image and an outline and keeps all data points within the specified distance of the outline (removes the "core" of the cell). It runs shrink_wrap and uses its results to do this.
- histogram2: This program creates a number of histograms of signal intensity (e.g., Calcium label) or volume as a function of distance of the object voxels from the nearest background voxel. If the "background" voxel represents the plasma membrane of a cell, the volume (or signal intensity, eg, Calcium) histogram this program produces describes, e.g., what percentage of the cell volume (or, eg, Calcium) is within 1 um of the plasma membrane. The volume histogram can be used to determine if intracellular objects are distributed randomly within the cell. Changes in the signal intensity histogram over time can indicate movement of signal (e.g., Calcium) within the cell.
- analyze_histo: Rather than apply a single threshold value to an entire image, this program produces a different threshold for each distance from the plasma membrane. It analyzes the histograms produced by histogram2 to calculate these thresholds. Different thresholds may be needed since signal intensity levels may vary as a function of distance from the plasma membrane and therefore noise levels (due to non-specific signal and/or Poisson noise) may also depend upon that distance.
- map_to_surface : This program takes a surface (which are those voxels which are nonzero) and one (or two) data images. It then maps (moves) the data voxels (nonzero voxels) to the position of the nearest surface voxel. It can be the first step in deciding (for example) whether cellular structures just below the plasma membrane are distributed randomly "on" the plasma membrane (a not infrequent question posed by physiologists).
- distance2surface_overtime.pl : Finds the distance (in pixels) of a specified point (over time) from the nearest pixel greater than 0 in a 3D image. Alternatively, it finds the distance to the boundary surface specified with the -surface option. Written to take a tracked gene in the nucleus and find its distance to the nuclear envelope over time.
Reference: Lifshitz L.M., Fogarty, K.E., Gauch, J M. & Moore, E.D.W. "Computer vision and graphics in fluorescence microscopy". Visualization in Biomedical Computing, SPIE vol. 1808, 521-534 (1992)
CellSim - Simulating chemical reactions and diffusion in cells.
CellSim is a program for 3-D reaction and diffusion simulation. Cellsim uses a Forward Time Centered Space fully explicit differencing scheme to solve the associated PDEs. Cellsim can run in parallel using MPI. Cellsim models multiple species of diffusible molecules and their reactions (e.g. ions and their binding targets). The simulation is saved as a series of images at specified time intervals. Images explicitly describing flow (since there can be large flow yet no net change in concentration) can also be saved.
References:
Kargacin, G., and F.S. Fay. "Ca2+ movement in smooth muscle cells studied with one- and two-dimensional diffusion model" Biophys. J. 60:1088-1100 1991.
Zou, H.L., M. Lifshitz, R.A. Tuft, K.E. Fogarty, and J.J. Singer "Imaging Ca2+ entering the cytoplasm through a single opening of a plasma membrane cation channel". J. Gen. Physiol. 114:575-588. 1999.
DAVE - Quantitative visualization of 3/4/5-D image data
The Data Visualization and Analysis Environment is a real-time interactive software system for displaying and analyzing multi-wavelength, 3-D, time-series images. DAVE simultaneously displays up to three image stacks, in red, green and blue. The images can be displayed as 2-D slices or full 3-D volumes, using several different methods for volume rendering. Surfaces (polygonal) or outlines (polyline) can be simultaneously displayed with volumes. Time series can be interactively reviewed in real-time, with selected time points held on screen for visual reference. Pixels or voxels of co-localization among the three images sets can be visually highlighted, segmented, and quantified. DAVE calculates percentage of overlap of regions of images, and the probability that such overlap could have occurred randomly. DAVE can render voxels as an individual cubes when the location or value of an individual voxel is desired. An "intelligent" 3-D cursor automatically snaps to the brightest nearby voxel when clicked. DAVE can remotely synchronize with another instance of DAVE, thus researchers in different locations see and interact with the same exact views. A somewhat outdated version of DAVE's help pages is online, as are some representative images and movies.
Reference: Lifshitz, L. M., Collins, J. A., Moore, E. D. W. & Gauch, J. "Computer vision and graphics in fluorescence microscopy". IEEE Workshop on Biomedical Image Analysis, Seattle, pp.166-175. 1994.
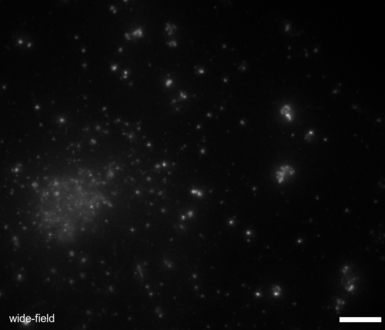
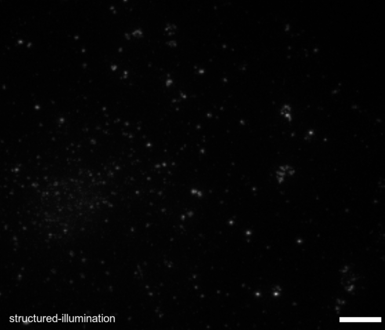
EPR - Image Restoration and super-resolution.
Over 20 years ago the BIG pioneered the development of image deconvolution for light microscopy and the resulting EPR algorithm 25 remains one of the best. EPR uses the light microscope's point spread function (PSF) in an iterative process to move out-of-focus light back to its originating position. Unlike many image restoration algorithms it is guaranteed to converge, and when given a finely-sampled point spread function it has been shown to increase lateral image resolution beyond the diffraction limit to less than 100 nm/pixel25. When combined with structured light microscopy (i.e. Structured Light Epifluorescence EPR or SLEEPR) it can achieve resolutions better than 50 nm laterally and 250 nm axially. EPR is computationally demanding. Recently Dr. Bellvé has produced both cluster-enabled and GPU versions of EPR, the latter of which can now restore images in seconds on a single workstation, make EPR for the first time amenable for High-Througput, high-speed and intelligent acquisition modalitis.
References:
Carrington W.A. and Fogarty K.E. “3-D Molecular distribution in living cells by deconvolution of optical sections using light microscopy.” In: Proc. of the 13th Annual Northeast Bioengineering Conference, K. Foster, ed., IEEE, pp108–111, 1987.
F.S. Fay, W. Carrington and K.E. Fogarty "Three-dimensional molecular distribution in single cells analysed using the digital imaging microscope." , J.Microscopy, Feb 1989.
Carrington WA1, Lynch RM, Moore ED, Isenberg G, Fogarty KE, Fay FS. "Superresolution three-dimensional images of fluorescence in cells with minimal light exposure." Science, 1995.
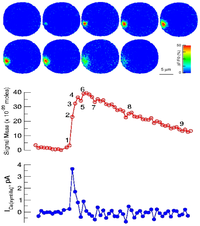
SignalMass - quantifying intracellular calcium microdomains.
SignalMass is program for the analysis of fast, time-lapse images of live cells loaded with a fluorescent calcium indicator for spatially and temporal local (microdomain) events (flashes of light) representing the release of calcium from internal stores through events called calcium "sparks" and "puffs".
During a Ca2+ "spark", free Ca2+ and Ca2+ bound to fluo-3 quickly diffuse away from the spark release site as Ca2+ continues to be discharged. To quantify the total fluorescence (and, ultimately the total Ca2+ discharged) arising from the binding of fluo-3 to the discharged Ca2+(i.e., the Ca2+ signal mass), the increase in fluo3/fluo4 fluorescence ) must be collected from a sufficiently large volume (and in 3D) to provide a measure of the total quantity of Ca2+ released.
References:
Zhuge, et al. J.Gen.Physiol. 2000
Zhuge, et al. Biophyscal J. 2006
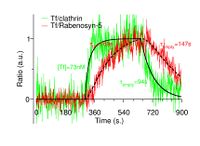
Kinetic Analysis of TIRF Time-Series Images
Among key endosomal components implicated in early endosome fusion are proteins such as early endosome antigen (EEA1), adaptor protein, phosphotyrosine interaction (APPL1), and Rabenosyn-5. To determine the role of endosomes containing these proteins, we imaged each one simultaneously with clathrin and Tf, using multi-color Total Internal Reflection Fluorescence Microscopy (TIRF-M). We developed an analysis regime for extracting the kinetics of the association and disassociation of Tf with these proteins over time.
In TIRF microscopy, the observed signal depends on both the intrinsic brightness of a fluorophore and its depth in the TIRF field. Images were processed to identify the positions (x,y,t) of all endosomes. At each endosome position the total fluorescence of the endosomal marker and of its cargo (e.g. Tf) were calculated. The fluorescence ratio of the cargo to its endosomal marker gives the relative amount of cargo independent of depth. Thus the amount of cargo associated with different endosomal markers (e.g. Rabenosyn-5) was tracked over time. For more details click Here
Reference: Navaroli et al. PNAS 2012
Generating Random Data/Simulations
- CellSim : Simulates chemical reactions and diffusion in 3D cells.
- connected_random_pts : Generates random points in a planar region, calculates how many connected components are created if a point is connected to all other points within a given radius.
- cross_sections_of_voronoi : Creates a random 3D voronoi tessellation and 2D image (cross-section) from it.
- ellipse : Creates an elliptical cell. It is used to model a squashed cell, and see how different it looks after blurring from a cylindrical cell.
- makebeads2 : Randomly distributes beads within a volume; created to simulate the distribution of glut4 labeled vesicles in the TIRF field of a microscope.
- oxygen : Calculates steady state partial oxygen pressure (P) as a function of distance from a blood vessel (r).
- random_paths.pl: Generates a random path (ie, diffusion of a single particle).
- vcspat : Calculates complete spatial randomness (Ripley's K function) of punctates in a volume.
- vcspat-models: Drops random objects in a volume. Objects can either be individual voxels, tiny images, or connected voxels in an image.
Statistics
- coloc3way image1[.i2i] image2[.i2i] image3[.i2i] [coloc-image[.i2i]] : Calculates two-way and three-way binary image colocation.
- overlap3: Produces a whole set of numbers with colocalization for each of many offset directions of one image relative to the other (to compensate for slight mis-registration between the images).
Finding events, Tracking
- exo6 : Identifies vesicle fusion events in a 2D time series. Intensity maxima are tracked, then each is analyzed to see if it meets the criteria to be a fusing vesicle.
- track : Tracks regions over time. Tracks written to stdout in rpts format. This is done by thresholding the input image. Resulting connected voxels define regions. Uses one of 3 tracking algorithms, lots of parameters can be specified to customize the tracking algorithm.
- trackinfo.pl : Analyzes output from track.
- track_pts : This programs takes a list of initial points in a 3D image and tracks them over time. It uses hill climbing or the brightest point in the neighborhood to link to in the next time point.
- paths4.pl : Reads tracking information from *.txt files, generates statistics for each track (speed, pathlength, distance, curvature). Statistics printed to stdout. There are lots of filtering and analysis options.
Miscellaneous
- parse_macros.pl (and associated .pl files) : Produces layered ImageJ macro files with variables passed through between them. This makes it much easier to write ImageJ macro programs which call other ImageJ macro programs. All of the parameter flags available in the called program are automatically propagated up to the calling program and made available to be set.
- diffusion2 : Uses VTK to diffuse (and visualize) random points in a volume.
- find_blobs : Uses maxima and zero crossings of the 2nd derivative to find blobs and their spatial extent; works in multiple resolutions.
- thin3Ds: This program is for thinning (i.e., finding the skeleton or medial axis) 3D objects. It is modified from Lee, Kashyap, and Chu (a parallel algorithm). thin3Dp is the same but in parallel.
- channel_current2 : Calculates the current which would be produced when each object in a Ryanodine labeled image releases calcium (e.g, a "spark") which activates nearby BK channels also labeled in the image (in a different wavelength).
- morph3d : Performes morphological processing on grayscale images.
- pwarpL : pwarp warps an image into a new image based upon a set of registered points from one image to the other. A line segment can also be specified as an old "point". The new point will match the best position along this line segment.
I2I - The BIG Image File Format
The I2I image file format dates back to the mid-1980s, built on a format first created by Dr. James Coggins.
The format is recognized by OME Bioformats and thus can be opened in OMERO, ImageJ and Fiji, as well as any other software incorporating Bioformats. Additionally, there is a I2I reader for Matlab and Scilab (available on request).
The basic format is a fixed size, 1024 byte, image header followed by the binary raster data. This allows the file data to be read sequentially and is therefore compatible with "pipes" where the output of one program becomes the input to the next without creating an intermediate file. Whenever possible, the BIG software allows piping of images following the Linux convention, i.e. the use of a "-" in place of an image file name redirects the input to stdin or output or stdout.
Ex.:
program1 input.i2i - | program2 - output.i2i
I2I image files have 4 dimensions, labled as X, Y, Z and N:
X, Y and Z are intended to represent spatial dimensions. N can be whatever the user wishes: time, channels, replicates, etc.
In practice, for images of 2-D over time, Z is considered time and N=1.
- The conventional file extension is .i2i
- The format was designed to be compatible with pipelines.
- The first 1024 bytes of the image file are the Image Header. The remaining bytes are the binary image pixel data, stored as a compact matrix ordered X by Y by Z by N, with X varying the fastest.
- The Image Header consists of two sub-sections
- The file organization section: 64 bytes
- The image history section(s): 15 x 64 bytes
- for a total of 1024 bytes.
The File Organization Section
Begins with 21 ASCII characters: Byte Offset Field Description ------ ----------- -------------------------------- 0 I|R|C Image data type (1 character) 1 <space> 2-6 xxxxx X dimension size (5 numeric characters, 0-9) 7 <space> 8-12 yyyyy Y dimension size (5 numeric characters, 0-9) 13 <space> 14-18 zzzzz Z dimension size (5 numeric characters, 0-9) *** See Note *** 19 <space> 20 B|L|<space> Data endian (1 character) IMAGE DATA TYPE A single ASCII character. Only supported file types are "I" 16-bit signed integer, "R" 32-bit floating point, "C" 32-bit complex (Real,Imaginary pairs) DIMENSIONS 5 ASCII numeric characters (0-9). Each of the X, Y, and Z Dimensions must be greater than or equal to one(1). DATA ENDIAN A single ASCII character. Increasing numeric significance with increasing memory addresses is known as little-endian. Only valid endian characters are: "B" for big endian (e.g. Java DataOutputStream) "L" or <space> for little endian (e.g Intel x86 including x86-64) A space is the same as an "L" for little endian. The next 10 bytes are binary and thus *** subject to endian interpretation *** Byte Offset Data Type Field Description ------ ------------ ------------ -------------------------------- 21,22 signed short Min Global intensity minimum (default 0) 23,24 signed short Max Global intensity maximum (default 0) 25,26 signed short X0 Image position in camera field: lower-left x-cooridinate (default 0) 27,28 signed short Y0 Image position in camera field: lower-left y-cooridinate (default 0) 29,30 signed short N Number of image sets in file *** See Note *** 31-63 Undefined and reserved for future use (default 0)
Image History Section(s)
The image history is comprised of 15 fixed-length, 64-character, ASCII lines,
beginning with an asterix ("*") followed by 63 alphanumeric (printable) characters.
The history lines are always sorted newest to oldest in a push-down fashion, discarding the oldest.
Each 64-character line must be padded with spaces to the end.
An undefined history line should be an asterix followed by 63 space characters.
Image File Size
The image Z dimension (byte offsets 14-18) is the total number of 2-D images in the file. The size in bytes of an I2I file is therefore 1024 + (X*Y*Z)*sizeof(type) HOWEVER For a 3-D image (X,Y,Z) the actual image Z dimension is Z/Nsets. Thus, the number of image sets N should always be > 0. However, for backwards compatability with old images, a value of 0 is interpreted as N=1.
Software Programs
The following is a description of many of the programs, roughly sorted by category.
Image-File-Type-Conversion
These programs convert mulitple image file formats into our I2I format.
alltiftoi2i
Repeatedly calls cstoi2i to convert 2D and 3D TIFF files to I2I format.
Syntax: alltiftoi2i [options] files...
Options: -h Prints this message.
-z n Number of planes (default is 1)
-u Data is 16-bit unsigned (see cstoi2i, note 2)
-2 Divide 16-bit data by two (same as -u)
-4 Divide 16-bit data by four
-8 Divide 16-bit data by eight
-v Verbose mode.
-D Debug mode.
allzeisstoi2i
Repeatedly calls zeisstoi2i to convert 2D and 3D TIFF files to I2I format.
Syntax: allzeisstoi2i [options] files...
Options: -h Prints this message.
-z n Number of planes (default is 1)
-u Data is 16-bit unsigned (see zeisstoi2i, note 2)
-2 Divide 16-bit data by two (same as -u)
-4 Divide 16-bit data by four
-8 Divide 16-bit data by eight
-v Verbose mode.
-D Debug mode.
Biorad3D
Syntax: % Biorad3d <rootname> nx ny nz channels timepoints
outputfile is 4-D image file named <root><channel>.i2i
Biorad3Db
Syntax: % Biorad3d <rootname> nx ny nz channels timepoints
outputfile is 4-D image file named <root><channel>.i2i
cstoi2i
Converts CELLscan or Metamorph(note 1) TIFF format images file to I2I format. Can be used for other TIFF formats with -z and -u options.
Syntax: % cstoi2i [options] [TIFF_file] [ < TIFF_file] [ > I2I_file]
TIFF_file TIFF format filename with extension I2I_file UMASS I2I format filename (must include .i2i) [ ... ] implies optional parameter. Input is from either filename or stdin(<). Output is only to stdout(>).
Options:
-h Prints this message. -z n Number of planes (default is 1) -u Data is 16-bit unsigned (note 2) -2 Divide 16-bit data by two (same as -u) -4 Divide 16-bit data by four -8 Divide 16-bit data by eight -v Verbose mode. -D Debug mode. -T Print ASCII text IFDs.
Notes:
1) Hacked (12/3/97) to determine the number of z-planes 2) Data is converted to 16-bit signed by dividing by two 3) Hacked to include comments 7/23/98 4) Hacked to better parse Metamorph comments 3/11/2002
Disclaimer:
This is not a completely robust TIFF implimentation and can fail if the IFDs are not ordered as expected. If the user fails to enter a comment, you will get a blank history line.
imformat
Formats a binary input image as .i2i file input pixels are 16-bit signed integers
Usage: imformat n x y z e <image >image
n is decimal number of bytes in header
x is decimal size of the X dimension
y is decimal size of the Y dimension
z is decimal size of the Z dimension
e is the data endian type {L or B}
Fixed on 5/9/2013 to handle truncated input files correctly
by filling out the missing pixels with zeros
imformatb
Formats a binary input image as .i2i file input pixels are 8-bit unsigned integers
Usage: imformat n x y z <image >image
n is decimal number of bytes in header x is decimal size of the X dimension y is decimal size of the Y dimension z is decimal size of the Z dimension
imformatR
Formats a binary input image as .i2i file input pixels are 16-bit signed integers
Usage: imformat n x y z e <image >image
n is decimal number of bytes in header
x is decimal size of the X dimension
y is decimal size of the Y dimension
z is decimal size of the Z dimension
e is the data endian type {L or B}
imformatrgb
Formats a binary input image as .i2i file input pixels are 8-bit unsigned integers
Usage: imformat n x y z <image >image
n is decimal number of bytes in header x is decimal size of the X dimension y is decimal size of the Y dimension z is decimal size of the Z dimension
putalltiftoi2i
Repeatedly calls cstoi2i to convert 2D and 3D TIFF files to I2I format.
Syntax: putalltiftoi2i dest [options] files...
Options: -h Prints this message.
-z n Number of planes (default is 1)
-u Data is 16-bit unsigned (see cstoi2i, note 2)
-2 Divide 16-bit data by two (same as -u)
-4 Divide 16-bit data by four
-8 Divide 16-bit data by eight
-v Verbose mode.
-D Debug mode.
rgbtifftoi2i
Syntax: rgbtifftoi2i newfilename[.i2i] red|green|blue file.TIF [file.TIF ...]
where: newfilename[.i2i] is the file name for the created i2i stack image red|green|blue is the color channel to use from the TIF files file[.TIF] is|are the sequence, in desired stack order, of the TIFF files
tifftoi2i
Syntax: tifftoi2i file1[.tif] [file2[.tif] ...]
Create I2I format image(s) from 16-bit 2-D TIFF. Image name(s) are the retained with .i2i extension.
xmgrtoi2i
Syntax: %xmgrtoi2i infile outfile infile xmgrace-style (& separated) data-set file name. outfile new image file name. Options: -size nx ny Size of image to create (required)
The xmgrace file is ACSII text. Data sets have one intensity value per line(record). Data sets are separated by "&". The image file has one set per row (x) starting at x=0. Each "&" starts a new row (y)
zeisstoi2i
Converts CELLscan or Metamorph(note 1) TIFF format images file to I2I format. Can be used for other TIFF formats with -z and -u options.
Syntax: % cstoi2i [options] [TIFF_file] [ < TIFF_file] [ > I2I_file]
TIFF_file TIFF format filename with extension I2I_file UMASS I2I format filename (must include .i2i) [ ... ] implies optional parameter. Input is from either filename or stdin(<). Output is only to stdout(>).
Options:
-h Prints this message. -z n Number of planes (default is 1) -u Data is 16-bit unsigned (note 2) -2 Divide 16-bit data by two (same as -u) -4 Divide 16-bit data by four -8 Divide 16-bit data by eight -X Divide 16-bit data by 256 (byte shift) -v Verbose mode. -D Debug mode. -T Print ASCII text IFDs.
Notes:
1) Hacked (12/3/97) to determine the number of z-planes 2) Data is converted to 16-bit signed by dividing by two 3) Hacked to include comments 7/23/98
Disclaimer:
This is not a completely robust TIFF implimentation and can fail if the IFDs are not ordered as expected. If the user fails to enter a comment, you will get a blank history line.
Image-Corrections
These programs perform various spatial and intensity-based manipulations.
autoscalexy
Autoscales each Z plane individually to 1 - 255
Usage: autoscalexy [options] image1 outimage options:
note: To view image use play. autoscalexy test - | play -
badpixel
Usage: % badpixel [options] inputfile outputfile
inputfile image file name
outputfile corrected image file name
Options:
Replace pixel (x,y) with
-a x y average of the 8 neighbors
-m x y median of the 8 neighbors
-p x y root of the sum of the squares of the 8 neighbors
-r x y square of the sum of the roots of the 8 neighbors
Replace each pixel of column (x,y1->y2) with
-A x y1 y2 average of the 6 neighbors
-M x y1 y2 median of the 6 neighbors
-P x y1 y2 root of the sum of the squares of the 6 neighbors
-R x y1 y2 square of the sum of the roots of the 6 neighbors
bgcor3d
bgsub3d Subtract Correct 3D (Z or time) bg from data
Input file 3D data image file
Output file The actual 3D data (above bg)
Options:
-Z n Z plane for BG
-E BG subtract each Z plane separately
-C x y Center of ROI
-R n Radius of ROI
-S n Number of sigmas(std.dev.) above mean bg (default=0)
-N sf Normalize by mean bg: image=sf*(image-bg)/bg (exclusive of -S)
-V value Subtract value from image (exclusive of other options)
-M n {file|-} Subtract the mode (histogram peak) of each Z plane (exclusive of other options).
n is stdev for histogram Gaussian smoothing (default=0, no smoothing)
file is filename for mode value(s), or - for stderr
bleachima
Usage: % bleachima [options] input_file output_file
Exponential bleaching correction for 2-D times series
Options:
-sets n Specifies the number of 3-D image sets
-A A0 A1 Single exponential: A0*exp(-z/A1)
-B B0 B1 Optional 2nd exponential: A0*exp(-z/A1)+B0*exp(-z/B1)
-C A0 A1 B1 (where B0 = 1 - A0)
-Z z1 z2 Fit z-planes z1-z2 with A0*exp(-z/A1)
Can be used repeatedly to specifiy disjoint sets of planes
(overrides -A and -B)
-R x1 x2 y1 y2
Restrict -Z to region (x1,y1) to (x2,y2)
-D n Image background value(baseline, def=0)
-V Verbose operations(def=not verbose)
flip
Flips image about X and/or Y, or Z axes
Usage: % flip [options] input_image output_image
where input_image Image file to flip output_image Flipped image file options: -sets n Specifies the number of 3-D image sets. -X Flip along X axis -Y Flip along Y axis -Z Flip along Z axis -T Transpose X and Y axes
Use flip -Y to display images right-side-up. Flipping modes are mutually exclusive.
imsets
Sets the number of images sets in an image file.
Usage: % imsets imagefile.i2i #
scratchng
Usage: % tranima [options] inputfile outputfile
Input file ScratchNG image file name
Output file black-level and gain corrected image file name.
Options:
-gains B4 B3 B2 B1 A4 A3 A2 A1
where the ports are labeled:
+-------------------+
2 | A4 | A3 | A2 | A1 |
|----+----+----+----|
1 | B4 | B3 | B2 | B1 |
+-------------------+
1 2 3 4
-nogains Sets all port gains to 1
Notes:
in-place corrections (12/05/2005)
default gains as of 8/10/2010:
1.000 0.947 0.942 0.966 1.018 1.055 1.035 1.024
replaces bad columns x=[160,481] with average of adjacent columns (3/24/2011)
updown3d
Syntax: updown3d file[.i2i] zsize [plateau]
file is name of source image file (.i2i is optional)
3D Deconvolution (EPR)
Exhaustive Photon Reassignment (EPR) is a method for Image Restoration, developed by the Biomedical Imaging Group at the University of Massachesetts Medical School. All versions of the program are based on the algorithm originally developed by Walter Carrington(1-3) with Kevin Fogarty (1-3) and Fay Fay (2,3). Basically, it is an iterative, least-squares reconstruction method with tikhonov regularization and a non-negativity constraint (no negative fluorescence). The micropscope (the forward problem) is modeled as a shift-invariant linear system (convolution).
The inputs are a series of optical section images of fluorescence acquired with a suitably equipped microscope (i.e. the data), a 3-D point spread function image, either theoretical or empirical (the PSF), and a "smoothness" parameter ("alpha"). The algorithm finds the object (true 3-D fluroescence distribution) that minimizes a weighted sum of the least-squares fit to the data and the "smoothness" (total energy) of the object. As the algorithm as constructed is minimizing a strictly convex function, it is garranteed to converge to a single global minimum (a unitque object).
The data optical sections do not have to be sampled on a regular grid, in x or y (laterally) or in z (axially). The object can be reconstructed on a finer grid than the data (sparsedata, binsample, superresolution-epr). When ER is combined with Structured Light epi-fluorescence microscopy (SLeEPR) it can achieve resolutions of 50nm in x,y and 150 nm in z.
(1) Carrington WA, Fogarty KE. 3D molecular distribution in living cells by deconvolution of optical sectioning using light microscopy.
In: Foster KR, editor. Proceedings of the Thirteenth Annual Northeast Bioengineering Conference. Vol. 1. New York: IEEE; 1987. pp. 108–110
(2) Three-dimensional molecular distribution in single cells analysed using the digital imaging microscope.
Fay FS, Carrington W, Fogarty KE. J Microsc. 1989 Feb;153(Pt 2):133-49
(3) Superresolution three-dimensional images of fluorescence in cells with minimal light exposure.
Carrington WA, Lynch RM, Moore ED, Isenberg G, Fogarty KE, Fay FS. Science. 1995 Jun 9;268(5216):1483-7
binsample
BINSAMPLE bins pixels n x,y and z while maintaining the original sampling Input file Image file name. Output file Image file name. Options: -bin x y z Specifies the spatial binning factor (default is 1)
Notes: The output image size is n-1 smaller where n is the binning factor for each axis Pixels are averaged when binned
epr_i2i
epr UMMC/BIG Jul 12 2012
NAME epr - Exhaustive Photon Replacement (EPR) restores contrast by removing residual out-of-focus light and improves resolution while maintaining numerical accuracy of 3-D images of specimens obtained with serial optical sectioning from wide-field or confocal light microscopy.
SYNOPSIS epr [options] before-image[.tif] after-image[.tif]
DESCRIPTION epr performs regularized, iterative image restoration with a non- negativity constraint. The before-image is a three dimensional (3-D) TIFF image composed of rectangular, regularly spaced, optical sections. Large images are decomposed into smaller, overlapping (in x and y only) image segments for restoration, and the restored segments are recomposed. The after-image is the restored 3-D image. All options must appear before the image file names, but the order of options is not important.
epr has the following options:
-h Prints basic help on syntax and available options.
Required Parameters
-psf file[.tif] The 3-D point spread function image file. The psf must
match the optical configuration used for the before-
image. The psf depth (number of z-planes) need only be
less-than or equal-to twice that of the before-image.
Extra z-planes are symmetrically discarded.
-smoothness alpha The alpha value where alpha is typically 0 < alpha < RNL.
-sm alpha RNL is the residuals noise limit as reported by prepdata
(see also). Smaller values of a correspond to less
smoothing. (RNL)^2 is usually a good starting choice for
alpha.
-iterations n Maximum number of iterations to perform. Iterating may
-it n terminate earlier if convergence is detected. (see
-convergence)
Control Parameters
-scaling n where n is after image scaling factor. Used to prevent
-sc n integer overflow when saving after image to file.
Default=1.0
-convergence n Criteria for terminating iteration, where n is << 1.
-co n 0.00001 represents true convergence. 0.001 usually
achieves 90-95% convergence in about half the number
of iterations. Values of n larger than 0.001 are not
generally recommended. Default=0.001
-axial n1 n2 The z-axis object extrapolation beyond the sectioned
-ax n1 n2 image data where n1 is before first plane and n2 is
after last plane. n is in planes. By default,
extrapolation extends +-1/2 the z-axis extent of the point
spread function image and should be sufficient. Smaller
values of n1 or n2 have the effect of spatially
constraining the restored object and should be applied
carefully.
-time n1 The timepoint where you want the restoration to occur (0 indexed) -t n1
-threads n The number of threads FFTW should use. Default=1 -th n
-transverse n The x-axis and y-axis object extrapolation in pixels.
-tr n The epr process extrapolates the restoration beyond the
bounds of each image segment in order to account for
exterior out-of-focus contributions. Default = 1/4 of X or Y
dimsenion PSF width, which ever is larger.
EXAMPLES To see the command line syntax
% epr
or
% epr -h
Given specimen image mycell_.i2i and point spread function image mypsf_.i2i
let smoothness equal 0.0005, maximum number of iteration equal 250
and convergence equal 0.001. Perform single-resoltion(default) EPR:
% epr -psf mypsf_ -smoothness 0.0005 -iterations 250 mycell_ mycell_r
FAQ (Frequently Asked Questions)
see http://...
FILES
xcomp:/home/epr/epr
xcomp:/home/epr/cs_epr.lo
SEE ALSO
prepdata
preppsf
Usage:
%epr [options] before-image after-image
For help on syntax and available optionos:
%epr -h
epr_gpu
epr UMMC/BIG Aug 8 2012
NAME epr - Exhaustive Photon Replacement (EPR) restores contrast by removing residual out-of-focus light and improves resolution while maintaining numerical accuracy of 3-D images of specimens obtained with serial optical sectioning from wide-field or confocal light microscopy.
SYNOPSIS epr [options] before-image[.tif] after-image[.tif]
DESCRIPTION epr performs regularized, iterative image restoration with a non- negativity constraint. The before-image is a three dimensional (3-D) TIFF image composed of rectangular, regularly spaced, optical sections. Large images are decomposed into smaller, overlapping (in x and y only) image segments for restoration, and the restored segments are recomposed. The after-image is the restored 3-D image. All options must appear before the image file names, but the order of options is not important.
epr has the following options:
-h Prints basic help on syntax and available options.
Required Parameters
-psf file[.tif] The 3-D point spread function image file. The psf must
match the optical configuration used for the before-
image. The psf depth (number of z-planes) need only be
less-than or equal-to twice that of the before-image.
Extra z-planes are symmetrically discarded.
-smoothness alpha The alpha value where alpha is typically 0 < alpha < RNL.
-sm alpha RNL is the residuals noise limit as reported by prepdata
(see also). Smaller values of a correspond to less
smoothing. (RNL)^2 is usually a good starting choice for
alpha.
-iterations n Maximum number of iterations to perform. Iterating may
-it n terminate earlier if convergence is detected. (see
-convergence)
Control Parameters
-scaling n where n is after image scaling factor. Used to prevent
-sc n integer overflow when saving after image to file.
Default=1.0
-convergence n Criteria for terminating iteration, where n is << 1.
-co n 0.00001 represents true convergence. 0.001 usually
achieves 90-95% convergence in about half the number
of iterations. Values of n larger than 0.001 are not
generally recommended. Default=0.001
-axial n1 n2 The z-axis object extrapolation beyond the sectioned
-ax n1 n2 image data where n1 is before first plane and n2 is
after last plane. n is in planes. By default,
extrapolation extends +-1/2 the z-axis extent of the point
spread function image and should be sufficient. Smaller
values of n1 or n2 have the effect of spatially
constraining the restored object and should be applied
carefully.
-time n1 The timepoint where you want the restoration to occur (0 indexed) -t n1
-cuda n1 The cuda device that you would like to use (1 indexed). -cu n1 The highest device number would be your display card.
FFTW will be used if a cuda device isn't specified or less than 1.
Restoration will fail if there isn't enough free memory on
your cuda device.
-threads n The number of threads FFTW should use. Default=1
Only applicable if using FFTW, and not CUDA.
-th n
-transverse n The x-axis and y-axis object extrapolation in pixels.
-tr n The epr process extrapolates the restoration beyond the
bounds of each image segment in order to account for
exterior out-of-focus contributions. Default = 1/4 of X or Y
dimsenion PSF width, which ever is larger.
EXAMPLES To see the command line syntax
% epr
or
% epr -h
Given specimen image mycell_.i2i and point spread function image mypsf_.i2i
let smoothness equal 0.0005, maximum number of iteration equal 250
and convergence equal 0.001. Perform single-resoltion(default) EPR:
% epr -psf mypsf_ -smoothness 0.0005 -iterations 250 mycell_ mycell_r
FAQ (Frequently Asked Questions)
see http://...
FILES
xcomp:/home/epr/epr
xcomp:/home/epr/cs_epr.lo
SEE ALSO
prepdata
preppsf
EPR
EPR is a shell script that checks to see if you are logged into either zirconium (the workstation in the analysis room) or barium (in Roughua's office). If yes, then it runs the CUDA version of epr using either the GTX 680 (zirconium) with 2Gb of memory, or the GTX 480 (barium) with 1.5 GB of memory, with options to show you iteration by iteration results. If you're using any other computer then it runs the Intel-cpu only version of epr. It passes the remainder of your command line on to the chosen epr program.
To see the help for the epr program itself
% EPR -h
Usual syntax:
% EPR -it 999 -sm # -co 0.003 -psf your_psf_file[.i2i] input[.i2i] output[.i2i]
Put 999 for iterations to make sure it doesn't stop too early. The smoothness value is whatever you would usually choose. I recommend using 0.003 for the convergence (rather than 0.001 or smaller) when using the NVIDIA version (zirconium or barium) as the lack of double-precision calculations can make it hard to squeeze the last bit out of the algorithm, and sometimes it will stop converging and you won't get an output image. If this happens anyway, then either try 0.005 for convergence and/or make the smoothness value a little larger, or ask Kevin for help.
If your image is really large then it might not fit into the memory of the NVIDIA systems. An image of 1300 x 1000 with fewer than 20 slices will work on zirconium (with 2GB), but to do more slices than this you will need to segment the image to something like 960 x 960 first (the reason has to do with allowable FFT sizes and the dimensions of the PSF, but this is a guideline).
You CAN to run more than one NVIDIA version of EPR at a time if there is enough available memory on the graphics board. Otherwise you'll get the error:
CUDA: out of memory error in file 'epr.cu' at line 1020
It's first-come first-serve.
linux_epr_fftw3_sse_z
epr UMMC/BIG 03/30/95
NAME epr - Exhaustive Photon Replacement (EPR) restores contrast by removing residual out-of-focus light and improves resolution while maintaining numerical accuracy of 3-D images of specimens obtained with serial optical sectioning from wide-field or confocal light microscopy.
SYNOPSIS epr [options] before-image[.tif] after-image[.tif]
DESCRIPTION epr performs regularized, iterative image restoration with a non- negativity constraint. The before-image is a three dimensional (3-D) TIFF image composed of rectangular, regularly spaced, optical sections. Large images are decomposed into smaller, overlapping (in x and y only) image segments for restoration, and the restored segments are recomposed. The after-image is the restored 3-D image. All options must appear before the image file names, but the order of options is not important.
epr has the following options:
-h Prints basic help on syntax and available options.
Required Parameters
-psf file[.tif] The 3-D point spread function image file. The psf must
match the optical configuration used for the before-
image. The psf depth (number of z-planes) need only be
less-than or equal-to twice that of the before-image.
Extra z-planes are symmetrically discarded.
-smoothness alpha The alpha value where alpha is typically 0 < alpha < RNL.
-sm alpha RNL is the residuals noise limit as reported by prepdata
(see also). Smaller values of a correspond to less
smoothing. (RNL)^2 is usually a good starting choice for
alpha.
-iterations n Maximum number of iterations to perform. Iterating may
-it n terminate earlier if convergence is detected. (see
-convergence)
Control Parameters
-scaling n where n is after image scaling factor. Used to prevent
-sc n integer overflow when saving after image to file.
Default=1.0
-convergence n Criteria for terminating iteration, where n is << 1.
-co n 0.00001 represents true convergence. 0.001 usually
achieves 90-95% convergence in about half the number
of iterations. Values of n larger than 0.001 are not
generally recommended. Default=0.001
-axial n1 n2 The z-axis object extrapolation beyond the sectioned
-ax n1 n2 image data where n1 is before first plane and n2 is
after last plane. n is in planes. By default,
extrapolation extends +-1/2 the z-axis extent of the point
spread function image and should be sufficient. Smaller
values of n1 or n2 have the effect of spatially
constraining the restored object and should be applied
carefully.
-time n1 The timepoint where you want the restoration to occur (0 indexed) -t n1
-threads n The number of threads FFTW should use. Default=1 -th n
-transverse n The x-axis and y-axis object extrapolation in pixels.
-tr n The epr process extrapolates the restoration beyond the
bounds of each image segment in order to account for
exterior out-of-focus contributions. Default=16 pixels
-OP wavelength NA n dx dz ideal
The optical configuration of the data and psf where
wavelength, dx and dz are in microns, n is one of
1.0 air
1.33 water
1.47 glycerin
1.515 oil
and ideal is < 1 for spherically aberrated systems.
-znear n The z-axis distance to first resolution reduction stage. -zn n Default=1.25 microns.
EXAMPLES To see the command line syntax
% epr
or
% epr -h
Given specimen image mycell_.i2i and point spread function image mypsf_.i2i
let smoothness equal 0.0005, maximum number of iteration equal 250
and convergence equal 0.001. Perform single-resoltion(default) EPR:
% epr -psf mypsf_ -smoothness 0.0005 -iterations 250 mycell_ mycell_r
FAQ (Frequently Asked Questions)
see http://...
FILES
xcomp:/home/epr/epr
xcomp:/home/epr/cs_epr.lo
SEE ALSO
prepdata
preppsf
linux_epr_pentium4
epr UMMC/BIG 03/30/95
NAME epr - Exhaustive Photon Replacement (EPR) restores contrast by removing residual out-of-focus light and improves resolution while maintaining numerical accuracy of 3-D images of specimens obtained with serial optical sectioning from wide-field or confocal light microscopy.
SYNOPSIS epr [options] before-image[.tif] after-image[.tif]
DESCRIPTION epr performs regularized, iterative image restoration with a non- negativity constraint. The before-image is a three dimensional (3-D) TIFF image composed of rectangular, regularly spaced, optical sections. Large images are decomposed into smaller, overlapping (in x and y only) image segments for restoration, and the restored segments are recomposed. The after-image is the restored 3-D image. All options must appear before the image file names, but the order of options is not important.
epr has the following options:
-h Prints basic help on syntax and available options.
Required Parameters
-psf file[.tif] The 3-D point spread function image file. The psf must
match the optical configuration used for the before-
image. The psf depth (number of z-planes) need only be
less-than or equal-to twice that of the before-image.
Extra z-planes are symmetrically discarded.
-smoothness alpha The alpha value where alpha is typically 0 < alpha < RNL.
-sm alpha RNL is the residuals noise limit as reported by prepdata
(see also). Smaller values of a correspond to less
smoothing. (RNL)^2 is usually a good starting choice for
alpha.
-iterations n Maximum number of iterations to perform. Iterating may
-it n terminate earlier if convergence is detected. (see
-convergence)
Control Parameters
-scaling n where n is after image scaling factor. Used to prevent
-sc n integer overflow when saving after image to file.
Default=1.0
-convergence n Criteria for terminating iteration, where n is << 1.
-co n 0.00001 represents true convergence. 0.001 usually
achieves 90-95% convergence in about half the number
of iterations. Values of n larger than 0.001 are not
generally recommended. Default=0.001
-axial n1 n2 The z-axis object extrapolation beyond the sectioned
-ax n1 n2 image data where n1 is before first plane and n2 is
after last plane. n is in planes. By default,
extrapolation extends +-1/2 the z-axis extent of the point
spread function image and should be sufficient. Smaller
values of n1 or n2 have the effect of spatially
constraining the restored object and should be applied
carefully.
-time n1 The timepoint where you want the restoration to occur (0 indexed) -t n1
-threads n The number of threads FFTW should use. Default=1 -th n
-transverse n The x-axis and y-axis object extrapolation in pixels.
-tr n The epr process extrapolates the restoration beyond the
bounds of each image segment in order to account for
exterior out-of-focus contributions. Default=16 pixels
-OP wavelength NA n dx dz ideal
The optical configuration of the data and psf where
wavelength, dx and dz are in microns, n is one of
1.0 air
1.33 water
1.47 glycerin
1.515 oil
and ideal is < 1 for spherically aberrated systems.
-znear n The z-axis distance to first resolution reduction stage. -zn n Default=1.25 microns.
EXAMPLES To see the command line syntax
% epr
or
% epr -h
Given specimen image mycell_.i2i and point spread function image mypsf_.i2i
let smoothness equal 0.0005, maximum number of iteration equal 250
and convergence equal 0.001. Perform single-resoltion(default) EPR:
% epr -psf mypsf_ -smoothness 0.0005 -iterations 250 mycell_ mycell_r
FAQ (Frequently Asked Questions)
see http://...
FILES
xcomp:/home/epr/epr
xcomp:/home/epr/cs_epr.lo
SEE ALSO
prepdata
preppsf
prepdata
prepdata UMMC/BIG 06/19/02
NAME prepdata - prepares a 2-D, 3-D or 4-D image data sets, acquired using a Digital Imaging (light) Microscope, for further processing (e.g. image restoration/3-D reconstruction)
SYNOPSIS prepdata [options] before-image after-image
DESCRIPTION prepdata performs all necessary corrections on a image data set based on the fluorescence D.I.M image formation/acquisition model, and can be used to perform basic imaging corrections to many forms of digitally acquired image data. The before-image argument is the name of a 3-D image set as acquired (DIM-1, DIM-2, CELLscan, UFM, or other). The after-image argument is the file name for the corrected image set, ready for further processing. The normal order of application of corrections (see -before) is basic[->background[->temporal]]. All calculations are performed using single-precision floating-point arithmetic. All options must precede input and output image file names.
Usage:
%prepdata [options] before-image after-image
For help on syntax and available options:
%prepdata -h
preppsf
preppsf UMMC/BIG 12/20/02
NAME preppsf - prepares a 3-D point spread function (psf) image data set, acquired using a Digital Imaging (light) Microscope, for use with image restoration/3-D reconstruction. Normally psf images are first processed using prepdata (see below) to apply basic corrections and any background corrections, but not temporal corrections.
SYNOPSIS preppsf [options] before-image after-image
DESCRIPTION preppsf performs further corrections on a psf image data set based on the fluorescence D.I.M image formation/acquisition model. The before- image argument is the name of a 3-D psf image set, acquired (DIM-1, DIM-2, CELLscan, UFM, or other) and processed using prepdata (see below) without the -norm option. The psf is extracted as a symmetric sub-region (square in XY) centered at the psf origin (-center), optionally normalized for constant total intensity (-norm), and optionally masked (-mask) to exclude any extramural data. The after-image argument is the file name for the processed psf image set. All options must precede input/output image files
Usage:
%preppsf [options] before-image after-image
For help on syntax and available optionos:
%preppsf -h
SNR
Usage: SNR [options] imagefile
imagefile data for SNR calculation
Options:
-gain n CCD gain (e/ADU)
-read n CCD readout noise (e RMS)
-bias n CCD bias value (ADUs)
-dark imagefile dark current image
-flat imagefile flatfield image
-offset dx dy offset of lower-left corner of data image
from lower-left corner of flatfield image
sparsedata
sparsedata UMMC/BIG 01/29/97
NAME
sparsedata - re-formats sparsely sampled images for EPR interpolation
and/or extrapolation
SYNOPSIS
sparsedata [-{h,sets,magnify|interpolate,extrapolate,z]
source-image[.i2i] destination-image[.i2i]
DESCRIPTION
The source image is re-formatted for EPR(see also) interpolation
and/or extrapolation by creating a "sparse" destination image sampled
on a finer(integer multiple), regularly-spaced spatial grid. Existing
data are placed in their corresponding sample positions. The
remaining grid positions are set to a unique value (-32768) that
indicates missing data to EPR. sparsedata is normally applied
following prepdata(see also) and before EPR. EPR must be provided
with a corresponding PSF (see note 1 below). For 4-D images,
re-formatting is repeated for each 3-D image set.
sparsedata has the following options:
-h Prints basic help on syntax and available
options.
-sets n Specify the number of 3-D image sets in
source-image. Overrides the sets number (if)
present in source-image and assigns the sets
number in new-image.
-magnify x y z Create intervening voxels between existing
-mag x y z voxels for each spatial axis. A value of 1
indicates no magnification for that axis, a
value of 2 doubles the sampling (equivalent to
-interpolate 1), etc.
-interpolate x y z Create intervening voxels between existing
-int x y z voxels for each spatial axis. A value of 0
indicates no interpolation for that axis, a
value of 1 create one intervening voxel
(equivalent to -magnify 2), etc.
-extrapolate x y z Create surrounding voxels outside the bounds of
-ext x y z the existing voxels for each spatial axis. The
new voxels are created symmetrically (+/-axis).
A value of 0 indicates no extrapolation for that
axis.
-zed file[.zed] Re-format the z-axis based on sampling protocol
described in the named file (see note 2 below).
Any z-axis interpolation (-interpolate or
-magnify) is applied AFTER -zed re-formatting
and cumulatively.
The -magnify and -interpolate options describe the same process and
are thus mutually exclusive. Use of both results in an error.
EXAMPLES
1) Interpolate one optical section between existing sections:
% sparsedata -int 0 0 1 in_image.i2i out_image.i2i
which is equivalent to doubling the magnification in z:
% sparsedata -mag 1 1 2 in_image.i2i out_image.i2i
2) Now also extrapolate four sections, two before and two after
the data:
% sparsedata -int 0 0 1 -ext 0 0 2 in_image.i2i out_image.i2i
3) Re-create a z-axis sampling protocol (7 optical sections at
-5,-3,-1,0,1,3,5) positioned so that -5 becomes the first
plane:
protocol.zed (see note 2 below)
+------------
| 6
| -5 1
| -3 1
| -1 1
| 0 1
| 1 1
| 3 1
| 5 1
+------------
% sparsedata -zed protocol.zed in_image.i2i out_image.i2i
The out_image has 11 optical sections (-5,-4,-3,...,5) .
4) Now also double the original z-axis spacing:
% sparsedata -zed protocol.zed -mag 1 1 2 in_image.i2i out_image.i2i
The out_image has 21 optical sections (-10,-9,-8,...,10).
FILES
SEE ALSO
prepdata
preppsf
epr
NOTES
1) The PSF must have the same sampling grid as new-image. However,
CCD detectors (generally) integrate over the entire area of a
pixel, as compared to optical sectioning, which measures at
discrete positions in z. The PSF in x and y must be sampled at the
new grid spacing, but the new pixels should be area integrations
corresponding to the original (source-image) pixel area.
2) The .zed files are simple text files describing the optical
sectioning protocol used for acquisition. They have the following
format:
The first line is the "origin" in z. This is added to all
following z positions to create an index(plane number)
greater than or equal to one.
Subsequent lines have two value separated by a (white)space:
the relative z position and the relative exposure time for
the corresponding image plane.
For 4-D images, the protocol is repeated for each 3-D image set.
3. A - in place of an image name means stdin or stdout.
superresolution-epr
Usage: superresolution-epr [-h|H] image[.i2i] psf[.i2i] smoothness XY-fold Z-fold
Description:
Superresolution EPR is a technique for restoring (deconvolving) an image using a finer pixel sampling
than the original image. This gets around the limitation on acheivable resolution imposed by the imaged pixel size.
Example: Given the command
$ superresolution-epr foo.i2i psf.i2i 1e-3 3 2
The program creates these files in your directory:
psf_3x3x2.i2i (interpolated psf.i2i) psf_3x3x2_sr.i2i (interpolated psf convolved with a 2-D square of the original pixel size) foo_3x3x2.i2i (foo.i2i resampled with NODATA (-32768) at new pixel positions) foo_3x3x2_sr1e-3.i2i (the restored image with the new pixel count and size)
The arguments XY-fold and Z-fold are the pixel sampling increase; e.g. a value of 2 means double the number of pixels in the restored image and half the pixel spatial size (double the sampling resolution). It is limited to 1, 2 or 3 in XY and Z for reasons of calculation time (proportional to fold increase) and storage space (it uses lots of random access memory).
Notes:
The program takes care of everything. You give it properly prepared image and psf files ready for regular EPR,
and it handles all the things need to make it work. I only recommend running this on large RAM machines and on images where the resulting superres (original size times the requested XY and Z fold increases) image will be under 1000x1000x100. And run it on the fastest CPU you can (that would be the new mizar and alcor).
Authors:
Karl D. Bellve <karl.bellve@umassmed.edu> Kevin E. Fogarty <kevin.fogarty@umassmed.edu> Walter A Carrington
References:
Superresolution three-dimensional images of fluorescence in cells with minimal light exposure. WA Carrington, RM Lynch, ED Moore, G Isenberg, KE Fogarty, and FS Fay Science 9 June 1995: 268 (5216), 1483-1487
Copyright:
© 2014 Biomedical Imaging Group, Univ. of MA Medical School, Worcester MA, USA
See also:
epr_i2i sparsedata preppsf preppdata
SLeEPR
Usage: superresolution-epr [-h|H] image[.i2i] psf[.i2i] smoothness XY-fold Z-fold
Description:
Superresolution EPR is a technique for restoring (deconvolving) an image using a finer pixel sampling
than the original image. This gets around the limitation on acheivable resolution imposed by the imaged pixel size.
Example: Given the command
$ superresolution-epr foo.i2i psf.i2i 1e-3 3 2
The program creates these files in your directory:
psf_3x3x2.i2i (interpolated psf.i2i) psf_3x3x2_sr.i2i (interpolated psf convolved with a 2-D square of the original pixel size) foo_3x3x2.i2i (foo.i2i resampled with NODATA (-32768) at new pixel positions) foo_3x3x2_sr1e-3.i2i (the restored image with the new pixel count and size)
The arguments XY-fold and Z-fold are the pixel sampling increase; e.g. a value of 2 means double the number of pixels in the restored image and half the pixel spatial size (double the sampling resolution). It is limited to 1, 2 or 3 in XY and Z for reasons of calculation time (proportional to fold increase) and storage space (it uses lots of random access memory).
Notes:
The program takes care of everything. You give it properly prepared image and psf files ready for regular EPR,
and it handles all the things need to make it work. I only recommend running this on large RAM machines and on images where the resulting superres (original size times the requested XY and Z fold increases) image will be under 1000x1000x100. And run it on the fastest CPU you can (that would be the new mizar and alcor).
Authors:
Karl D. Bellve <karl.bellve@umassmed.edu> Kevin E. Fogarty <kevin.fogarty@umassmed.edu> Walter A Carrington
References:
Superresolution three-dimensional images of fluorescence in cells with minimal light exposure. WA Carrington, RM Lynch, ED Moore, G Isenberg, KE Fogarty, and FS Fay Science 9 June 1995: 268 (5216), 1483-1487
Copyright:
© 2014 Biomedical Imaging Group, Univ. of MA Medical School, Worcester MA, USA
See also:
epr_i2i sparsedata preppsf preppdata
Convolution
The prgrams are useful for fast linear filtering, and for creating simulated data.
blur2d
Usage: blur2d [options] image psf result
Image files (do not use any file extension): image .i2i file image to blur psf .i2i file image to serve as psf result .i2i file blurred image
Option(s):
-S n scale factor for result (default=1)
n<1 prevents integer overflows
n>1 minimizes integer truncation
-N x y dimensions of convolution space
(default dimensions are image + 1/2 psf)
-M x y dimensions of output image
(defaults are dimensions of input image)
-Z pad image convolution space with zeros
(default is pad image with edge values)
-X cross-correlation instead of convolution
-xnorm xcorr with normalization: I=(I-Imean)/Imean
implies -X -Z -D but not -0
-D Do not normalize the psf
-0 No DC component in psf
-V verbose mode (default is not verbose)
blur2d will choose a convolution space size large enough to prevent wrap-around. This size must also 1) be even in the X axis, and 2) have largest prime factors < 19. The image is padded with the edge pixel values to avoid sudden transitions to zero, except with the -Z option.
blur3d
Blurs an image by a psf. FFTs are done with 4 byte floats, so total space needed will be 4 times the padded image size (in voxels) plus the size of the two original images (image and psf) The input image can be real or short int format. The psf must be short int format. note: performs a convolution (not a correlation). note: fixed a small bug. 10/17/11
Usage: blur3d [options] image psf newimage
options:
-S #: scale (multiply) output values by # (to prevent underflow or overflow)
voxels which after scaling are > 32767 are set to 32767. voxels < -32766 are set to -32766.
-p # # #: pad the image in x, y, and z by the specified amounts
by default image is padded (with 0) by 1/4 psf width all around,
so its size for blurring becomes bigger by psf/2.
psf is always padded to the same size the image becomes.
Padding is stripped off before newimage is written, so it will be the same size as
the original image.
-N # # #: pad so that the image has the specified dimensions after padding (just prior to FFT).
-n #: pad with # instead of boundary pixel values.
-Z: pad with zero instead of the boundary pixel values.
-c # # #: the center of the psf is at (#,#,#) zero indexed.
the default is to take the brightest pixel as its center.
-C: don't center the psf at all (default is to center on the max, unless -c is specified).
-P: normalize the psf so each slice (after padding) averages to 1. -P acts like an optical PSF.
the default is to normalize so the entire psf (after padding) has an average pixel value of 1
(the default keeps total light constant).
-d: don't normalize at all.
-o: do a cross correlation instead of a blur (convolution)
-R: output image as 4 byte floating point instead of short integer.
-v: verbose. print more info to stderr.
note: entire image name (with extension) is NOT required. note: can read from stdin or write to stdout (use a dash instead of the file name) note: a compressed image will be automatically be uncompressed.
smooth2d
smooth2d 2D image smoothing
Input file The image to be filtered.
Output file The filtered image.
Options:
-3 3x3 approx to gaussian filter: [[1,2,1][2,4,2][1,2,1]]
-B n NxN box filter
-T n NxN triangle filter
-G n Gaussian kernel where n = std.dev.
-IO Nin Nout
Difference of average of inner Nin x Nin and outer Nout x Nout regions
-RMS n calculate local RMS value over n x n pixels
-S n image scale factor.
-X 1D X-axis(horizontal) smoothing.
-Y 1D Y-axis(vertical) smoothing.
-pos Positive-quad-only box filter
-v|-V Verbose
smooth3d
smooth3D 3D image smoothing
Input file The image to be filtered.
Output file The filtered image.
Options:
-3 3x3x3 gaussian approximation.
-G sd1 sd2 p sd3 Gaussian kernel where sd = std.dev. in pixels
sd1 is x,y
sd2 is z rising phase
p is z duration of peak (usually zero)
sd3 is z falling phase
-Z Z-axis only filtering (xx,y extent=0)
-S n image scale factor.
Note: The half-amplitude point of a gaussian occurs at 1.177*sd
Modeling/Simulation
These programs are used when modeling images of cells.
checknoise
Usage: checknoise [options] imagefile [textfile] Imagefile An image file name (extension optional) Options: -Z n -N n -M
COSINEex
Usage: % scaleima [options] input_file output_file
Options: -size x y (pixels) -period n (pixels) -phase n (degrees) default=0
Notes:
makebead
MAKEBEAD Make a 3D fluorescent bead model Output file The converted image file. Options: -dim x y z Image dimensions x,y, and z (pixels) -D n n bead inner and outer diameters (microns) -R n inner rise std.dev. (microns) -F n outer fall std.dev. (microns) -S n n n X,Y,Z pixel dimensions (microns) -X n number subpixels per pixel
makeDOG
Usage: % makeGaussian [options] output_file
Options: -DOG s1 s2 Difference-of-Gaussians sigmas (pixels) s1<s2 -scale n Gaussian unit area scale factor (default is +1.0)
makeImpulse
Usage: % makeImpulse [options] output_file
Options: -dim nx ny nz image size (pixels) -orig x0 y0 z0 impulse location(pixels) -scale n impulse scale factor (default=1)
noise
Abstract:
This program takes an image (ADUs), adds a background (ADUs),
multiplies by a scale factor (-s), and adds photon (Poisson) and read (Gaussian) noise appropriate to a given detector (-c|-e,-g)
Usage: noise [options] inp_image out_image
Options:
-c RCA DIM-1 type of camera -c TI DIM-2 type of camers -c LL128 SCRATCH type of camera -c PM512 RATCH type of camera -c LLraw SCRATCH type of camera (input in electrons) -c photons input in photons(just adds shot noise) -e e_ADU electrons per A to D -g gain gain -n sigma read noise as electrons RMS -b number value of background (added to input, not output) -s number value of scale_factor (applied to input after -b)
Image files (do not use any file extension):
inp_image .i2i input file of image data out_image .i2i output file of image data
NOTE:
'-' in place of an image name means stdin or stdout.
psf3di
Computes a 3-D microscope point spread function image.
Syntax: % psf3di [options] output_image_file
Options: -help Extended abstract explaining method -verbose Show wavefront data -size x y z Pixel sizes (microns) -dim x y z Psf extents (pixels) -lambda n Wavelength in vacuum (microns) -NA n Numerical Aperture. of objective lens -sqrt Save square-root of intensities (more dynamic range)
-nominal n0 z n1 thickness n2 -actual n0 z n1 thickness n2
-cglass z z-plane poistion of the cover glass (default is (zdim+1)/2)
Nominal refers to the nominal objective lens corrections.
Actual refers to the actual imaging parameters.
Where:
n0 n(D) of object medium
(nominally same as n2)
z Distance(microns) from cover glass
(nominally 0)
n1 n(D) of cover glass
(1.522 +/- 0.004 is standard)
thickness Thickness(microns) of cover glass
(170 +/- 10 is standard no. 1)
n2 n(D) of lens immersion medium
(air=1.0, water=1.33, oil=1.515)
Example: objective is a Zeiss 63x 1.4NA Planapo % psf3di -NA 1.4 -nominal 1.515 0 1.522 170 1.515 ...
Image-Reformatting
These programs are used when changing the pixel sampling of images.
chres
chres changes image resolution(trilinear interpolation) Input file image file Output file image file Options: -S n n n X Y and Z voxel scale factors -E Operate on each Z plane separately
concateima
Usage: % concateima [options] image image... output_image
Concatenates image files together
Options: none
hugesegz
Syntax: hugesegz inputfle[.i2i] outputfile[.i2i] zstart zend
matrixima
Usage: % matrixima [-lead string] [-tail string] -row ... -col ... output_image
Assembles multiple time-course well images into a matrix that can be viewd with play
where the canonical well image name is [{lead}]{row}{col}_{position}_{objective}_{filter}[{tail}].i2i
required: -row n str [str [str...]] typically B, C, D, etc. -col n str [str [str...]] typically 02, 03, 04, etc.
important(check defaults): -pos[ition] n position in well: 0 <= n <= 99 (default is 0) -obj[jective] n object lense: 0=5x, 1=10x, 2=20x (default is 1) -fil[ter] n filter position: 0=phase, 1=GFP, 2=RFP (default is 0)
optional: -lead string filename leading string (default is none) -tail string filename trailing string (default is none)
-bin n bins pixels nxn (default is larger of rows and cols) -h[elp] additional information and examples.
Notes: The first well (row 1, col 1) image MUST exist. Skipped wells (missing image file) will be included in matrix but left blank.
Colocalization
These prgrams calculate measures of colocalization between/among images..
coloc3way
coloc3way image1[.i2i] image2[.i2i] image3[.i2i] [coloc-image[.i2i]] Two-way and three-way binary image colocation Options: -T n1 n2 n3 image1|2|3 Thresholds(>) default: -T 0 0 0 -E operate on Each image plane separately (2-D mode)
Meaning of output column headers: 1=>2 percentage of image-1 pixels found with image-2 1=>23 percentage of image-1 pixels found with both image-2 and image-3 1=>!23 percentage of image-1 pixels found with image-3 but not with image-2 1=>2!3 percentage of image-1 pixels found with image-2 but not with image-3
Object-Segmentation
The programs perform various types of segmentations/classifications of pixels into objects, etc.
addlines
Adds polyline|pixel objects to image files
Usage: % addlines image textfile [options] image image input image file textfile polyline|pixel object description file image output image file Options: -O label n select labeled object with w=n(superseeds -P) -P n default w for all objects -C n mark centroids with w=n(superseeds -P) -T truncate floating point coordinates to integer (default is to round) -0 use zero-indexed coordinates (default is one-indexed) -silent redirect stderr to /dev/null Defaults: all objects selected w = -1 centroids not marked
Object description file format: 1)file is ASCII byte stream 2)blank lines are ignored 3)anything from a # to end-of-line is a comment 4)objects begin with the object= key and continue until the next object or the end-of-file 5)x,y,z coordinates are the position of the pixel 6)w coordinate is the value assigned to the pixel(s)
Legend:
| indicates a choice of parameters
[ ] indicates an optional parameter
{ } indicates a set of number separated by spaces
do NOT include these symbols in the data
Object description format:
object=name # name may include spaces
[closed] # ensures closure of a polyline object
# applies only to ordered boundaries
[z=n] # default z when none specified in coordinate
[everyplane] # apply as 2-D(x,y) coordinate list to every z-plane in image
[w=n] # set pixel intensities to w(superseeds command line options)
[centroid=x y] # 2-D centroid position of closed boundary
boundary=ordered|unordered|chain-code [xy|xyz|xyw|xyzw]
[{x y}|{x y z}|{x y w}|{x y z w}] # one pixel/line
[{x y}|{x y z}|{x y w}|{x y z w}] # repeated...
[break] # next pixel begins disjoint curve
# not compatable with closed option
[{x y}|{x y z}|{x y w}|{x y z w}] # repeated...
countobjs
Usage:
countobjs [options] outputfile image1.i2i [image2.i2i]
Description:
Count objects in co-localized images. If only one image given the objects will be counted in that one image. Copious amounts of information will be written to the output file. A - for outputfile will write to stdout. outpufile can be analyzed with objs2bb.pl and objs2dist.pl
Options:
-C # # : set opacity and brightness level to these values (0-255 only) -k : use original images for data analysis (so scale and black level not important and
-t option can be in same units as original, unscaled, image values)
-S # # # : image # scale and black level (default autoscale image)
-r # # # # # # : region of image to count
(-r x0 y0 z0 x1 y1 z1)
( ^lower left ^upper right)
(zero indexed coordinates)
(default whole image)
-s # # : size of smallest and largest objects to count
(default 1-100000)
-m # # : min and max iod of allowable objects (applied to image1 only).
-t thresh1 thresh2 : thresholds (applied after -S or default scaling, unless -k specified).
Like left brightness slider in DAVE.
if only one image, second number still required, but ignored.
-o <image.i2i> : output image with all objects represented by the voxel
with the maximum intensity in the object
the intensity of the voxel will be the IOD of the object.
-O <image.i2i> : output image with all pixels in each object having
the intensity of the original objects IOD.
NOTE: this image will be in floating point format.
use float2int to convert it back to "normal" format.
-Q <image.i2i> : output image with all pixels in each object having
a unique id # (1-# of objects). Must be < 32766 objects.
NOTE: this image will be in standard signed short int format.
-M # : When countobjs is given 2 images, this flag will
turn of some subset of the voxels for colcalization.
calculations.
0: turn off colcalized voxels (default on)
1: turn off image 1 voxels (default on)
2: turn off image 2 voxels (default on)
-q : quiet. don't print out all objects, but just summary statistics
-Z : simple analysis, no sliders, opacity,etc. can use with -t option.
-h : show this message.
-H : show even more help.
Notes:
A - in place of an image name means stdin or stdout. Source code in /storage/big1/lml/jac/was_invitro/Projects/countobjs
DoTheTrackingAndRatios
Syntax: DoTheAnalysis.csh [--random] rootfilename wavelength threshold(edges) [threshold(newprog1)|.]
Finds all local 2-D maxima above 1st threshold(edges) in 491|561
Computes the vesicle ratio of TFIs of 660/{491|561} in a 5x5 box centered at each maximum
Options:
--random randomize the 660 location realive to the 491|561 Set the 2nd threshold (newprog) to a period(.) for automatic denominator TFI thresholding
newprog1
foo image1[.i2i] image2[.i2i] image3[.i2i] textfile
Two-way intensity-based vesicle colocalization
image1 primary image (denominator)
image2 secondary image (numerator)
image3 image of vesicle IDs (2-D maxima)
Options:
-IO inner outer +-size (pixels) of region for local average intensiy calculation
default: 2 3 (i.e. 5x5 and 7x7)
-random n randomly shift vesicle paths by adding uniform random variable [0..n] pixels
default: 0
-T threshold primary (denom.) image local average intensity must be > threshold
(default 0)
Notes: 8/2/2010 output to textfile includes (ix,iy) of those positions passing threshold test
trackves
trackves 2D vescile tracking Inputfile_1 Original vesicle time series Inputfile_2 Filtered vesicle time series Outputfile Tracked vesicles time series. Options: -T vesicle intensity threshold (> or =) for filtered image (default 1) -id n vesicle ID modulo n (default 256) -xy n search n pixels in x and y to link vesicle path (default 1) -z n search n pixels in time (z) to link vesicle path (default 1) -dur mark vesicels with duration rather than ID -crop n crop the filtered image n pixels from edges (default 0) Notes: Only single paths allowed (no branching, first come first servered) setting -z > 1 no longer guaranties FCFS matching and branching may occur
planimeter3
PLANIMETER3: Measure length and area of closed contours Usage:
planimeter3 [options]
Options:
-I input_file Image file name(s). A maximum of three
images can be entered. The first one is displayed
as green, the second as red and the third as blue.
Voxels colocalised between any two images are shown
as white
-R rpts file. If it exists on input, it is read in and
contours contained in it are displayed and can be deleted
or edited. If it does not exist, it will be saved on
exit unless Quit is pressed first
-S n n n Image number, display scale and black-level -X n n range of X planes (low,high) -Y n n range of Y planes (low,high) -Z n n range of Z planes (low,high) -M n Magnification (zoom)
This program allows you to draw boundaries around a cell or object The boundaries can be drawn in any order and can be erased if required.
Use: Left mouse button: move between planes (as in play). Middle mouse button: draw boundary.
Once a object has been defined pressing the middle mouse button will start drawing a contour for that object, or, if the contour has already been started, continue it. Each subsequent perimeter is associated with the previously defined object. To change the object or start a new object use the pop-up menu.
Backspace - undraw previous point (may be repeated).
Keypad + : Zoom in. Keypad - : Zoom out.
l - (toggle) show n planes in addition to the current plane.
Where n is 3 (default) but can be set from 1 - 9 by pressing the appropriate numerical key. Pressing the l key again will show just the current plane
v - (toggle) while drawing will result in a vertical line being
drawn from the current position when the mouse is moved.
h - (toggle) while drawing will result in a horizontal line being
drawn from the current position when the mouse is moved.
Control (toggle) - Toggle on and off scaling for the images.
Notes: A maximum of 20 separate objects can be defined for
contouring. Each object is represented by a different
colour.
The only limit on the number of contours that may
added to an object is the number of Z planes.
There is a limit of 4096 points per drawn contour,
however there is no limit on the number of points
of a contour in a file that is read-in
pMapping
Maps (postive only) image intensities into probability P that pixel is from G1 rather than G0.
Usage:
% pMapping [-{G|B|S|P|Q}] input_file[.i2i] output_file[.i2i]
Options: -G1 A0 x0 A1 x1 s0 Double-Gaussian parameters (floats) -G2 A0 x0 A1 x1 s0 s1 Double-Gaussian parameters (floats) -G ... Same as -G1 (for backwards compatability) -B n Camera bias level (integer, default=0) -S n Image scale factor (float, default=100) -P Make P image (default) -Q Make (1-P) image instead
Notes:
Image pixel intensities (x) are modeled as belonging to one of two Gaussian probabilty distributions (G):
-G1: G0=A0*exp(-(x-x0)^2/(2*s0^2)) and G1=A1*exp(-(x-x1)^2/(2*(x1/x0)*s0^2))
-G2: or G1=A1*exp(-(x-x1)^2/(2*s1^2))
where x1 > ix
If x>0 then the probability that pixel belongs to G1 is: P=G1/(G0+G1)
and the probability it belongs to G0 is: Q=(1-P)
Else x<=0 and P=Q=-32768 (no data)
repeat_rpts
Syntax: % repeat_rpts filename count
Filename is the .rpts file to reproduce Count is the number of Z planes to reproduce
Output is to stdout
Example:
% repeat_rpts one.rpts 10 >many.rpts
Utilities
The programs perform various useful functions.
cull
Choose and order columns of text
Usage: cull [-{h,c,a,s,n,1,2,...}
Options:
-h print this message -c ignore comments (from # to end of line) -a ignore ampersands (from & to end of line) -s suppress blank output lines -n number rows -1,-2,... next column to print
hist
This program prints image history records.
Usage: hist imagefile
hist2list
hist2list filenames
Prints history information for .i2i files
NOTE: works works with wildcards
NOTE: works with .Z files
immax
INFOCUS Find position of brightest pixel in a 3D data set Input file The 3D image file. Options:
infocus
INFOCUS Find most infocus Z plane of 3D data set Input file The 3D image file. Options: -X output infocus X position -Y output infocus Y position -Z output infocus Z position (default) -N normalize z-planes (MGL=1) Output is a number (Z coordinate) sent to stdout
printpixels
Syntax: printpixels [options] imagefile [ > textfile ]
Options:
-X x1 x2 keep pixel coordinates between x1 and x2 inclusive
-Y y1 y2 keep pixel coordinates between y1 and y2 inclusive
-Z z1 z2 keep pixel coordinates between z1 and z2 inclusive
-low n keep pixels valued > low
-high n keep pixels valued < or = high
-silent suppress output of non-warnings/errors to stderr
Output is to "stdout"
stats
Usage % stats [-h] <infile >outfile Options -h types this message Computes minimum, maximum, mean and standard deviation of a string of numbers read from stdin.
imsets
Sets the number of images sets in an image file.
Usage: % imsets imagefile.i2i #
Image-Visualization
DAVE
Data Analysis and Visualization Environment is our primary 3D visualization tool for 3-D, 4-D and 5-D images (x,y,z,time,color).
NAME
dave - Data Analysis and Visualization Environment.
This program reads in rpts files in the format put out by
planimeter and displays a wire frame or surface model of the outlines.
Typically use -n 0 -u -kptcolor options with rpts paths which are not closed outlines.
It also reads in image files (-I option) and displays them.
-H will print more help.
USAGE:
dave [options] rptsfile1 rptsfile2 ...
OPTIONS:
-z # : magnification to apply in z (default = 3.000000)
Zspacing relative to xy spacing. Must be an integer.
-I name : image file (.i2i) of voxel data. You do not need to include
the ".i2i" in the name. Also, DAVE can handle compressed images.
You need a -I in front of each image name. If two images are
specified the first will be shown in green and the second in red.
All image files must be the same dimensions.
If the image name is iargs, all arguments without an option
flag are considered to be images
-S # # # : Image number (1 indexed), display scale, and black-level.
(If Image number = all apply scale and black-level to all images.)
By default, DAVE rescales each image file linearly so that the
brightest voxel maps to 255 and the dimmest voxel maps to 0.
The mapping is o = (i-black)/scale
where i is the input value and o is the output value and
"black" is set to the image minimum and "scale" to (max-min)/255.
This option lets you change the scale and black level. Any data
values which map above 255 get set to 255, those mapping below 0
get set to 0. This is the same a "play".
Sometimes an image has a few very bright pixels which cause the
max to be large; the large scale factor then makes most of the
image look dim. This option provides a way around that.
-S all 1 0 -S 1 4.2 2 will rescale all images with scale =1, black =0 except image 1
(the -S all option must be specified prior to the scalings of other specific images)
-Srange start stop scale black: apply to the range of images (1-indexed, inclusive).
-s : rescale each image z plane separately (with its own scale and
black level)
-x #1 #2 #3 : Use the image histogram to pick image rescaling values.
(If Image number = all apply values to all images.)
This is a way of trying to get around the above problem automatically.
Instead of using the brightest and dimest voxels to determine
black level and glScalef(see -S above, $2, $3) look at the entire histogram
of voxel values. The voxel value below which #2 percent of the
image values fall will be considered the image "minimum".
Similarly, the voxel value above which #3 percent of the values
lie will be considered the image "maximum". Rescaling is then
performed o = 255*(i-min)/(max-min), where o values
above 255 are set to 255 and o values below 0 are set to 0.
#2 should be based upon the percentage of voxels you expect to
be background voxels. #3 is the percent of voxels you want to
map to full white. Typical reasoning might be as follows:
If your image has equal number of voxels in all intensity levels
then you might want 1/255 of the voxels to end up background and
1/255 to end up full white, thus #2 and #3 might be about .4 .
The rescaling is applied to image (#1) (1 or 2).
-t : keep original image around for calculating image statistics.
By default, only the remapped values (0-255) are used.
This takes more space.
-T # dir : Transpose image # in some direction (dir can be XY, XZ or YZ).
Transpose takes place prior to z magnification.
-nogui : start dave with main rendering window only.
-noborder : rendering windows have no border.
-slideshow #: start dave with slide show.
-contents : show hypertext help for dave via netscape
-h : print this help message.
-H : print long form of help message (including examples).
-K : single-set texture model (1/3 space when in 2d texture mode, but limits sideways viewing).
-ft : use extra memory to make changing time points (-4) fast in texture mode, fast textures.
uses 2x the number of bytes of the original 4D image additional space w/out compression,
about 1/2 the number of bytes with compression.
-nocomp : don't use texture compression (4x more space than with compression,
perhaps better image quality)
Notes:
Much more extensive help is given when dave is running;
click on the HELP button in the DAVE Master widget.
EXAMPLES:
-------------------------------------------------------------
To read in an image file whose z spacing is 250 nm and xy spacing 125 nm:
dave -z 2 -K -I image.i2i
To read in two image files, specifying a scale and black level for the first
and using a histogram rescaling for the second, z spacing already equal to xy:
dave -z 1 -I image.i2i -I image2.i2i -S 1 10 15.3 -x 2 5 1
To read in three image files (see the Series menu item on the Stuff menu):
dave -I image.i2i -I image2.i2i -I image3.i2i
To read in an image and paths from an rpts file:
dave -I image.i2i -n 0 -u -kptcolor paths.rpts
To read in a 4D image (-tdim only needed if image header not set correctly):
dave -4 -tdim 20 -I image.i2i
For DAVE's extensive help pages Click Here
For some images created with DAVE Click Here
montage
Abstract: Displays a montage of 2-D images from a 3|4-D image.
Syntax: montage input_file Input file 3|4-D image file name.
Options:
-S n n Image display scale and black-level
-E Scale each Z plane individually
-Z n n 1st and last Z planes to display
-I n Increment between Z planes
-M n Magnification (zoom)
-R n The number of rows of images in montage
-C n The number of columns of images in montage
-NB Display window without a border
-o x y Display window at screen postion (x,y)
-false Display data using B-C-G-Y-R falsecolor
-frame Add a white frame around each image
-up Display consecutive images bottom-to-top
-across Display consecutive images left-to-right (DEFAULT)
-updown2one sets
Add a white frame representing up/down sampling
play
Usage: % play [options...] file [file2 [file3] ]
file(s) 2D, 3D or 4D image file name (with or without .i2i). FILES MUST BE OF SAME DIMENSIONS!
- Accepts input from stdin("<") or pipe("|") and titles as "<stdin>"
Options:
-sets n Number of 3-D image sets (default=1)
-S n n Image display scale and black-level
-M n Magnification (default=1)
-P n c Map intensity n to color c:{N R Y G C B M W}
-Z n n Range of Z planes (low, high)
-L Limit playback at range of Z planes
-U image values > black-level are displayed > 0
-B n n Scale bar length(microns) and microns/pixel
-project Display maximum intensity projection of Z-planes
DATA/ANALYSIS
-IO inner outer
Set the pixel readout (MIDDLE mouse button) ROI size where:
inner is the radius (+/- center pixel) of the additive ROI (default=0)
outer is the radius (+/- center pixel) of the subtractive ROI (default=0)
-name string
"string" replaces image filename (window title) as root name for creating files
DATA/PSF PREPARATIONS
-psf wavelength NA nD size spaced mask
Visualize the psf extent as a green circle
(wavelength, size and spaced must be in microns)
See preppsf for corresponding options
-min n Displays pixels valued less than "n" as blue
(default is -nomin)
-nomin Disables minimum function
-max n Displays pixels valued greater than or equal to "n" as red
(default is -max 32767)
-nomax Disables maximum function
-nonneg same as -min 0
ANIMATION
-movie {f|r|b}
Sets movie running forward, backword or both
-speed n Sets movie speed n={1,2,....9} (default=2)
-master Makes this a syncronous master
-n n Master(*) divides frame no. by n
-slave Makes this a syncronous slave
-pipe Both a master and a slave
-color Pseudocolor scale (default is gray scale)
-timebar Display time-bar instead of frame number
-hide Do not display the frame number
-o x y Screen origin (x,y) for window
-sched z n
Pause n video fields WHILE displaying plane z
-labels file
file contains one or more lines of x,y,z and labels
formatted |xxxxx yyyyy zzzzz string....
LEFT MOUSE left/right manual movie control MIDDLE MOUSE readout pixel position and value RIGHT MOUSE pops up Main menu
To adjust contrast/brightness (replaces SGI edmap program)`: choose Main menu => Color menu => Help for built-in edmap this will print instructions in the terminal window
To use Z/Time Plot Analysis: A-Key then Hold(!) down the MIDDLE MOUSE button, or press the SHIFT and A-Keys together If you use the first method to enter: release the MIDDLE button to exit If you use the second method to enter: press the A-Key again to exit
To use ROI Analysis: choose Main menu => ROI Analysis (or press the t-key on the keyboard) LEFT MOUSE size or move ROI box MIDDLE MOUSE left/right changes threshold
To use Intensity linescan: choose Main menu => Intensity linescan (or press the l-key on the keyboard) LEFT MOUSE draw scan line through image
To save the current image display(s) as 24-bit color, portable anymap files (.ppm): choose Main menu => Save image(s) => Current image (or > key on keyboard) or choose Main menu => Save image(s) => Entire image set (or = key on keyboard)
To save the entire image set as an MPEG movie:
choose Main menu => Save image(s) => Create MPEG (.mpg) movie from PPM set {+}
or press + key on keyboard. The movie speed will be the same as the current play speed)
To save the entire image set as an Quicktime movie:
choose Main menu => Save image(s) => Create Quicktime (.mov) movie from PPM set {-}
or press - key on keyboard. The movie speed will be the same as the current play speed)
To syncronize 2 movies:
% play -master master.i2i | play -slave slave.i2i
New features:
* "p" key toggles maximum-intensity projection on|off (see -project command line option)
* contrast widget includes multiple colors (beyond gray) for (mono)chroma
* on-image labels are available (see -labels) and can be toggled off|on using the "shift+L" key
* "}" key increases stereo parallax. "{" decreases parallax. Default parallax is 0 (no stereo).
* cycle among multiple images (file1, file2, file3) using END key
play3d
Usage: % play [options...] file
file 2D, 3D or 4D image file name (with or without .i2i).
- Accepts input from stdin("<") or pipe("|") and titles as "<stdin>"
Options:
-sets n Number of 3-D image sets (default=1)
-S n n Image display scale and black-level
-M n Magnification (default=1)
-P n c Map intensity n to color c:{N R Y G C B M W}
-Z n n Range of Z planes (low, high)
-L Limit playback at range of Z planes
-U image values > black-level are displayed > 0
-B n n Scale bar length(microns) and microns/pixel
-project Display maximum intensity projection of Z-planes
DATA/ANALYSIS
-IO inner outer
Set the pixel readout (MIDDLE mouse button) ROI size where:
inner is the radius (+/- center pixel) of the additive ROI (default=0)
outer is the radius (+/- center pixel) of the subtractive ROI (default=0)
-name string
"string" replaces image filename (window title) as root name for creating files
DATA/PSF PREPARATIONS
-psf wavelength NA nD size spaced mask
Visualize the psf extent as a green circle
(wavelength, size and spaced must be in microns)
See preppsf for corresponding options
-min n Displays pixels valued less than "n" as blue
(default is -nomin)
-nomin Disables minimum function
-max n Displays pixels valued greater than or equal to "n" as red
(default is -max 32767)
-nomax Disables maximum function
-nonneg same as -min 0
ANIMATION
-movie {f|r|b}
Sets movie running forward, backword or both
-speed n Sets movie speed n={1,2,....9} (default=2)
-master Makes this a syncronous master
-n n Master(*) divides frame no. by n
-slave Makes this a syncronous slave
-pipe Both a master and a slave
-color Pseudocolor scale (default is gray scale)
-timebar Display time-bar instead of frame number
-hide Do not display the frame number
-o x y Screen origin (x,y) for window
-sched z n
Pause n video fields WHILE displaying plane z
-labels file
file contains one or more lines of x,y,z and labels
formatted |xxxxx yyyyy zzzzz string....
LEFT MOUSE left/right manual movie control MIDDLE MOUSE readout pixel position and value RIGHT MOUSE pops up Main menu
To adjust contrast/brightness (replaces SGI edmap program)`: choose Main menu => Color menu => Help for built-in edmap this will print instructions in the terminal window
To use Z/Time Plot Analysis: A-Key then Hold(!) down the MIDDLE MOUSE button, or press the SHIFT and A-Keys together If you use the first method to enter: release the MIDDLE button to exit If you use the second method to enter: press the A-Key again to exit
To use ROI Analysis: choose Main menu => ROI Analysis (or press the t-key on the keyboard) LEFT MOUSE size or move ROI box MIDDLE MOUSE left/right changes threshold
To use Intensity linescan: choose Main menu => Intensity linescan (or press the l-key on the keyboard) LEFT MOUSE draw scan line through image
To save the current image display(s) as 24-bit color, portable anymap files (.ppm): choose Main menu => Save image(s) => Current image (or > key on keyboard) or choose Main menu => Save image(s) => Entire image set (or = key on keyboard)
To save the entire image set as an MPEG movie: choose Main menu => Save image(s) => Create MPEG movie from PPM set (or + key on keyboard) (note the MPEG speed will be the same as the current play movie speed)
To syncronize 2 movies:
% play -master master.i2i | play -slave slave.i2i
New features: 1) "p" key toggles maximum-intensity projection on|off (see -project command line option) 2) contrast widget includes multiple colors (beyond gray) for (mono)chroma 3) on-image labels are available (see -labels) and can be toggled off|on using the "shift+L" key 4) "]" key increases stereo parallax. "[" decreases parallax. Default parallax is 0 (no stereo).
rgbmerge
Takes one to three 2D grayscale images and merges them into a singel RGB image.'
Syntax:
rgbmerge R|G|B:filename.ext [R|G|B:filename.ext [R|G|B:filename.ext] ] merged.ext
Where:
R:, G:, or B: indicate the desired color channel for that image. Omitted color channels are black by default. The case of the color channel (R|r, G|g, B|g) does not matter. The order of the images on the command line does not matter.
Examples:
Merge a green JPEG image and a blue PNG image into one color TIFF image rgbmerge G:one.jpg B:two.png result.tif is the same as rgbmerge b:two.png g:one.jpg result.tif
Valid image file types are those recognized by ImageMagick.
Image-Processing
A collection of various and sundry programs for image processing.
Like many of the programs listed abouve, these routines are designed to work with image piplines. Substituting a dash "-" for an image name routes input from <stdin> and/or output to <stdout> Routines can be chained together with the comandline pipe character "|" for easy prototyping of complex functions.
----
derivima
derivima [-{diff|lr|exp,scale,max}] inputfile outputfile
dFoverF
Compute a fluorescence time-ratio image.
Usage:
dFoverF [-{h,sets,norm,range,noratio,AC,median,threshold,all,mask,scale,(no)flags}] input_file output_file
Use -h to get further help
diffima
derivima [-diff f|c|b] inputfile outputfile
dilateima
Usage: % dilateima [options] input_file output_file
Performs 2D dilation of given pixel value. Pixels not dilated are left intact.
Options: -P n Pixel value to dilate (def=1) -A also dilate all pixels > n
edges
Copyright 2010 University of Massachusetts Medical School and the Biomedical Imaging Group All rights reserved. Explicit permission to use this program must be received prior to use. Revised - May, 2014
Syntax: edges [options] input_file output_file
input_file The image to be filtered.
output_file The filtered image.
Options:
-LAP x y Laplacian kernel
size in x (pixels)
size in y (pixels)
-DOG s l Difference of gaussians kernel:
std.dev. of smaller gaussian (pixels)
std.dev. of larger gaussian (pixels)
-E n Extent (+/-no. of sd's) of DOG kernel
-M n image mean to preserve [0,1]
-S n image scale factor.
-I invert filtered image
-V verbose
erodeima
Usage: % erodeima [options] input_file output_file
Performs 2D erosion of given pixel value.
Options: -P n Pixel value to erode (def=1) -F n Value to fill eroded pixels (def=0)
FRETratio
FRETratio file [threshold [dx dy] ]
the threshold is applied to the YFP(upper) image. default is 0. the shifts (dx,dy) are applied to the CFP(lower) image. defaults are 0 and 0.
histima
Usage: histima [options] imagefile >textfile
Imagefile An image file name (extension optional)
Options:
-bins n Number of bins in histogram (default=256)
-size n Number of grey levels per bin
(default = max-intensity / number-of-bins)
-bg n Value for lowest bin (default=0)
Notes:
Histogram is output to stdout as ASCII compatable
with xmgr. (see /usr/local/xmgr)
Example:
% histima -bins 100 my_image | xmgr -pipe &
kinetic-analysis
Syntax: kinetic-analysis numerator[.i2i] denominator[.i2i] filtered-denominator[.i2i] threshold results[.txt]
or kinetic-analysis help
where:
numerator the ratio numerator (dependent) time series image denominator the ratio denominator (independent) time series image filtered-denominator corresponding image used for identifying 2-D maxima > threshold for ratio calculation threshold 2-D maxima intensity threshold (> or =) applied to filtered-denominator image results name of a text file for the kinetic time course results (see help)
help provides more information
lsr_align
Least Square Residual based image alignment.
Usage: LSRalign [options] in-image out-image in-image images to align out-image aligned images Options: -sets n Number of 3-D images sets -range dx dy +-range to align -first always align to 1st Z plane
maskedsmul
maskedsmul rescales image intensities in outlined regions
Usage: % maskedsmul {options] image1 image2 image3
image1 image to correct
image2 image with mask outline
image3 output image
Options:
-I n scale factor for pixels inside mask (def=1)
-O n scale factor for pixels outside mask (def=1)
-P n value for boundary pixels (def= -1)
-silent redirects stderr to /dev/null
Notes:
1) The outline pixels are considered outside the line
2) Use planimeter and addlines to create mask image
maski2i
Usage: maski2i [options] image1 outimage options:
-T # <= # = 0, > # = 1 [Default = 0]
OR -C # Complement, >= # = 0, < # = 1 [Default = 0]
mathi2i
mathi2i (Sep 20, 3013)
Basic pixel operations (+,-,*,/,max,min) between images
Syntax:
mathi2i [options] input-image-name input-image-name output-image-name
Options:
-a|s|m|d Performs image addtion, subtraction, multiplication or division respectively
-max|min Minimum or maximum of the two values at each pixel
-and|or Boolean operation: zero -> 0, non-zero -> 1, result is always 0 or 1
-S n1 n2 Scale(multiply) 1st and 2nd images by n before performing operation
Notes:
All calculations are done as floating point. Output images is integer truncated to [-32767:+32767].
If either image image value is -32768 ("no data") then output is always -32768.
Division by zero is undefined and result is -32768 ("no data") at that pixel.
(you can make "no data" pixels show up as red(R) with the command: play -P -32768 R ...)
maxima
Usage: % maxima [options] input_file output_file
Identifies 3D image maxima (peaks). The all non-maximal pixels are set to zero, unless the -F options is used. Then, non-maximal pixels are preserved and maximal pixels are set to the specified value.
Options: -T n maximum threshold value n -7 maximum if > 7 of 8 neighbors -E define maximum as > or = (mutally exclusive with -7) -F n flag maximum pixels with value n -Z z-axis maximum only -R input image is 4-byte floating point
If the output is omitted, the output is an ASCII list of maxima positions as (x,y,z,intensity)i sent to stdout.
normbg
Usage: % tranima [options] inputfile outputfile Input file image file name Output file image file name. Options: -roi xleft ybottom xright ytop -smooth nplanes (+/-planes to smooth, default=0) -zref zplane (default=1) -skip zstop zstart (repeat as needed) -corrbg (correct planes for dark errors, default is no)
nv-coloc-analysis
Syntax: nv-coloc-anal.csh results[.txt] folder1 [folder2 [ folder3 [...]]]
1) All the folders named on the command line will be analyzed. 2) Results of the colocalization analysis are appended to the named results textfile.
If the results textfile does not exists it will be created.
openclose2d
openclose 2D gray scale erosion(min) and dilation(max) Input file The image to be filtered. Output file The filtered image. Options: -open r erosion->dialtion over radius r (pixels) -close r dilation->erosion over radius r (pixels)
OTFz
Syntax: OTFz psf[.i2i] z-spacing(um) wavelength(um) NA(objective) nD bead(um)
OTFx
Syntax: OTFx psf[.i2i] pixel-size(um) wavelength(um) NA(objective) bead(um)
OTFy
Syntax: OTFx psf[.i2i] pixel-size(um) wavelength(um) NA(objective) bead(um)
padima
Usage: % padima [options] input_file output_file
Pads an image (on both sides) in X, Y and/or Z
Options: -B n Extent of padding in XY -X n Extent of padding in X -Y n Extent of padding in Y -Z n Extent of padding in Z -V n Value of padding pixels (def=0)
reduceima
REDUCEIMA reduces image resolution and size by binning Input file Image file name. Output file Image file name. Options: -sets n Specifies the number of 3-D image sets. set the binning factors: start(1 is first pixel) end(-1 is last pixel) incr(-1 is last-first+1) -X n n n X axis start, end and increment in pixels -Y n n n Y axis start, end and increment in pixels -bin bx by Equivalent to: -X 1 -1 bx -Y 1 -1 by -Z n n n Z axis start, end and increment in pixels -P Same as -Z 1 -1 -1 -N n n n Time axis start, end and increment in pixels -S Eliminate axes of size 1 on output set the binning mode -T Keep total(sum) of pixels -A Keep average of pixels -L Keep lowest(minimum) pixel -H Keep highest(maximum) pixel (DEFAULT) -M Keep median (not mean!) pixel -when Keep Z position of lowest|highest|mean pixel (combine with -L|-H|-M)
radcolpsf
RADCOLPSF: Radially averages psf images Input file An image file to be radially averaged. Output file The averaged image file. Options: -C n n Center of rotation (x,y)
ratio2wl
syntax: ratio2wl fura[.i2i] threshold(WL-1) [dark[.i2i]]
register
Usage: register [options] in-image out-image in-image images to register out-image registered images Options: -SSC Stochastic Sign Change method (default) -range dx dy +-range to register -first always register to 1st Z plane
RITS-fusion-analysis
Syntax:
RITS-fusion-analysis filename-root event_no.
RITS-process
This is the main application to fit RITS fusion model to TIRF time-series images
Syntax: RITS-process RFP[.i2i] GFP[.i2i] rootname
1) the coordinate textfile must be rootname.txt 2) images are named rootname_r|g_###.i2i 3) individual output graphs are named rootname_###.ps 4) the files of collected graphs are named rootname.ps and rootname.pdf
Necessary files:
RITS-select-event RITS-fusion-analysis RITS-2D-fusion-analysis-exe RITS-2Dto3D-fusion-analysis-exe RITS-fusion-analysis.par
RITS-select-event
RITS-select-event RFP[.i2i] GFP[.i2i] x0 y0 radius z0 z-before z-after rootname id
rotateima
Usage: % rotateima input_image outputimage Input file 3D An image file to be rotated. Output file 3D The rotated image file. Options: -F file File containing 3D transformation matrix -C n n n Center of rotation (x,y,z). -S n n n Scaling applied to X, Y, and Z axes. -R n n n Rotation about X, Y, and Z axes (degrees). -P n Perspective (distance to eye in pixels) -D nx ny nz Fix the output image size as nx,ny,nz
-arrow x0 y0 x1 y1
Defines an arrow from tail (x0,y0) to tip (x1,y1).
Image is rotated within the XY plane such that the
arrow is pointing up (increasing Y).
rotproj
ROTATEIMA: Produces a sequence of 3D image rotations and
projections of varying image attributes
Input file An image file to be rotated.
Output file The rotated image file.
Options:
-M n Projection mode: n=1 voxel SUM,
n=2 voxel MAX,
n=3 SUM w/opacity.
n=4 RANGE shading
-K n Optical Density factor: OD = n * intensity
(when mode=3) opacity = OD/(1+OD)
-T n Intensity threshold
-I c Interpolation N|L (Near-neighbor | Linear)
-C n n n Center of rotation (x,y,z).
-S n n n Scaling applied to X, Y, and Z axes.
-R n n n Rotation about X, Y, and Z axes (degrees).
-P n Perspective (distance to eye in pixels)
-V n n Viewport size (X, Y)
-Z n n Z-dimension clipping planes (zmin,zmax)
-A n n n Start, stop, delta rotation angles (deg)
-F Flag pixels outside data bounds with -32768
scaleima
Usage: % scaleima [options] input_file output_file
Options:
-Z z1 z2 dz scale Re-scale intensities of Z-planes
z1 to z2 stepping dz by scale
-V Verify -- list scale factors
Notes: Multiple Z ranges may be specified. Z ranges may be specified in arbitrary order. Any Z-plane not specified is scale by one(1.0) Maximum of 32767 Z-planes allowed.
segment
Usage: % segment [options] input_image output_image
Options:
-sets n Specifies the number of 3-D image sets.
-X n n n X axis start, end and increment in pixels
-Y n n n Y axis start, end and increment in pixels
-Z n n n Z axis start, end and increment in pixels
-N n n n Image set start, end and increment in pixels
-I n Include n'th Z plane in repeat series given by -Z option
(planes in repeat series are number from 1 to z-increment)
-ROI x0 y0 x1 y1 Corners of a rectangular region to segment
(alternative to -X and -Y options with increments of one)
-flag n Flag padding pixels with intensity n (default = -32768)
-S Eliminate axes of size 1 on output
Notes: -I option can be used to reorder planes in series.
If the region of interest extends outside of XY plane, the segmented image is padded with the flag value(-flag)
signalmass
During a Ca2+ "spark", free Ca2+ (diffusion coefficient, D = 250 μm2/s) and Ca2+ bound to fluo-3 (D = 22 μm2/s; Smith et al. 1998) quickly diffuse away from the spark release site as Ca2+ continues to be discharged. To quantify the total fluorescence arising from the binding of fluo-3 to the discharged Ca2+(i.e., the Ca2+ signal mass), the increase in fluo3/fluo4 fluorescence ) must be collected from a sufficiently large volume to provide a measure of the total quantity of Ca2+ released. Custom software was used to process the images and extract signal mass information for each spark event. The signal mass time course for each spark was computed from the two-dimensional, widefield fluorescence images of fluo-3 according to the following equations.
Total fluorescence,
Ftotal(t)= SUM[F(x+∆x, y + ∆y, t)].
The fluorescence F is summed over a 13.7-μm square region (41 pixels on a side in the x-y plane) surrounding the spark epicenter pixel (x,y) as determined from the ΔF /F0 images from Formula 1.
Signal mass,
sm(t) = G*(Ftotal(t)-Ftotal(t0))
The signal mass sm(t) is the change in total fluorescence FT(t) over the baseline fluorescence FT(t0) times the detector gain G (see below). The time t0 corresponds to the image immediately preceding the beginning of the spark. The beginning of the spark event was identified as the first image having a flux, i.e., an increase in total fluorescence (FT) relative to the preceding image, exceeding 2 SDs of the noise(1).
(1) Dynamics of signaling between Ca(2+) sparks and Ca(2+)- activated K(+) channels studied with a novel image-based method for direct intracellular measurement of ryanodine receptor Ca(2+) current.
ZhuGe R, Fogarty KE, Tuft RA, Lifshitz LM, Sayar K, Walsh JV Jr. J Gen Physiol. 2000 Dec;116(6):845-64.
Syntax:
signalmass radius [SF] < listfile > outfile
where:
radius is the halfedge size of bounding box SF is the [optional] scale factor for scalnge the dF/Fo traces (default=1) listfile is the spark|puff|syntilla description textfile |filename x y z | x y z |filename x y z | x y z outfile is the table of signalmass statistics
SLMrecon
SLMrecon phase0[.i2i] phase120[.i2i] phase240[.i2i] output-image[.i2i] Structure illumination optical sectioning according to T.Wilson Options: -S n SL reconstruction image scale factor (default: 1.0) -WF filename[.i2i] Save sum of three phases as WF image
SLMreconS.csh
syntax: SLMreconS root{-0.i2i,-1.i2i,-2.i2i}
root-0 is phase 0, root-1 is phase 120, root-2 is phase 240
spectrum
Usage: % spectrum [options] input_file output_file
Computes 2D FFT of image. The output file is complex FFT (.cxi file) unless -M or -E option used(.i2i file).
Options:
-A Apodize image with raised cosine window
-Z Pad image with zeros (default)
-P Pad image with mean-gray-level
-M Compute magnitude spectrum(not -E)
-E Compute energy spectrum(not -M)
-C Center spectrum at N/2(with -M or -E)
-LOG Log transform of magnitude or energy spectrum
-DC Zero the DC component of FFT
-N x y FFT size (Nx by Ny)
-S x y Pixel size in X and Y(def=1,1)
-F image Filter input_file using image as Fourier mask
-xlines dist order(s) width
Filter input_file using X layer lines where:
dist is no. pixels to layer line
orders are the line(s) to include
terminated with a zero.
width is of number of lines about this line
-ylines dist order(s) width
Filter input_file using Y layer lines
-R x y Filter out components within radius n of DC
Notes:
-A option scales spectrum by 1/mean-squared-value of
apodization window function
-S option scales spectrum by pixel area
-F Alternative to creating a convolution kernel
sqrtima
sqrtima [-S n] inputfile outputfile
statima
Copyright 2010 University of Massachusetts Medical School and the Biomedical Imaging Group All rights reserved. Explicit permission to use this program must be received prior to use.
Usage: statima [options] imagefile [textfile]
Imagefile An image file name (extension optional)
Textfile Optionally write statistics to file
Options:
-sets n Specifies the number of 3-D image sets.
-X n n left and right X coord for Region-Of-Interest
-Y n n bottom and top Y coord for R.O.I.
-Z n Statistics on single Z plane = n
-E Statistics on each Z plane
-P Mean and variance of positive(>0) pixels.
Sets threshold equivalent to -T 0
-T n Pixels with intensities less than or equal (< or =) n are ignored
Default is no threshold unless -P is invoked (see above)
Combine with -P to change threshold
-SE Standard error instead of standard deviation
-S Show image statistics (default)
-H Show image gray level histogram in portrait format (default)
Combine -H -S in this order to show both statistics and histogram
-L Show image gray level histogram in landscape format (not implemented)
-silent Redirects sdterr to /dev/null
Example: % statima -X 101 150 -Y 41 70 -T 10 -E -S my_image shows just the image statistics of those pixels greater than ten(10) in a 50 by 30 pixel rectangular region, Z-plane by Zplane, in the image in file my_image.i2i
thresholdima
Biomedical Imaging Group University of MA Medical School Copyright 2010
Syntax: %thresholdima [options] input_file output_file
input_file Image file name.
output_file Image file name.
Options:
-T n Threshold (<) intensity (default=0)
-V n Replacement value (default=0)
-S Subtract rather than threshold
-R low mid high
Triangular ramp function
low < low -> 0
mid = mid -> mid
high > high -> 0
-P n Power of ramp function (default=1)
TIRFex
Usage: % scaleima [options] input_file output_file
Options: -z planes the mean excitation distance for TIRF (default=0) -e fraction the fraction of maximal excitation due to epi background (default=0) -zref plane the z-plane where the relative TIRF excitation equals 1.0 (default=0)
Notes:
TIRF_WF
TIRF_WF [options, -h] inputfile outputfile
tranima
Usage: % tranima [options] inputfile outputfile
Input file Image file name.
Output file Translated image file name.
Options:
-T x y z Translation in X, Y and Z
(Positive translation is right, up, rear)
-rotate Translations are circular (DEFAULT)
- what goes off the edge wraps back on the opposite edge.
-shift Translations are not circular
- what goes off the edge is lost
- image is padded with -32768
Translations are rounded to the nearest integer value.
transp
Usage: % transp [option] inimage outimage Options: -sets n Number of image sets (4-D: default=1) -XY Transpose X and Y axes -XZ Transpose X and Z axes -YZ Transpose Y and Z axes
Signal-Processing
These programs work in the time domain on simple 1-D or 2-D (x|t,y) data
digfilt
NAME
digfilt - abstract
SYNOPSIS
digfilt [options] <stdin >stdout
DESCRIPTION
digfilt...
digfilt has the following options:
-lowpass Hz -3dB frequency
-highpass Hz -3dB frequency
-timestep seconds Sampling interval
EXAMPLES
SEE ALSO
NOTES
CAVEATS
LAST REVISION
--/--/--
BUGS
pdcr
Prints out columns of file Usage: pdcr [options] {file|-} options:
-c n # of columns -p n # of points to print out
straighten
Computes average pixel intensity along a single curve traced using planimeter program by mapping pixels to closest(euclidian distance) point along the line. Only points within a given distance from the line (-s option) are used and ONLY positive pixels are mapped.
Usage:
% straighten [options] image rptsfile [image]
where
image input image file name
rptsfile points along cell center line (planimeter)
image optional output image file name where output
image pixels contain average intensity at
position on line to which pixel was mapped
(ALL pixels are mapped for this operation)
Options:
-S n Maximum dist from centerline (def=nearest)
-N Normalize the center position [0..1]
-Z n Do only Z plane = n
-P Output image(optional) pixels replaced with
line position instead of average intensity
-A n Normalize average along line to n
-J Replace pixel intensities in output image
JUST under the curve.
-NR Do not rewind points file each time
-V Verbose mode
-xmgr Xmgr compatable output
-neg Allow negative values
line position (x) and average intensity (y) are sent to stdout and can be piped to graph|plot or Xmgr
Examples:
% straighten -S 10 image1 image1.rpts | graph | plot plots the average of those pixels 10 pixels or less from the centerline given in image1.rpts
% straighten -S 10 image1 image1.rpts > image1.graph sends the output to the file named image1.graph
% straighten -S 10 image1 image1.rpts image2 creates the pixel classification map instead
Simulation
analyze_simulation.pl
This program takes one or more outputs of a simulation (e.g. output of run1 or run2)
For each z slice (time point) in each output: blurs it with a psf, pulls out the in-focus slice.
Then calculates total concentration CHANGE (relative to time 0) as a function of distance
from the calcium entry site (assumed to be the brightest point over all time after blurring).
This is written to a file with the same root as the input simulation, but with a .xy extension.
This can be viewed with xmgrace, or read into matlab (readxmgr) and visualized with acurves().
Assumes simulation was at 100 nm resolution. Uses a fixed psf with 300x300x150 nm voxels.
Several scale factors are applied, so comparisons of magnitudes between files are not valid
(but comparisons between time pts from the same file are fine).
analyze_simulation.pl [options] sim.i2i [additional input files]
options:
-zrange=#:#:# startz stopz dz, inclusive, zero indexed. default is all z slices (i.e., all time pts).
-verbose print out extra into to stderr as it goes along
-keepfiles keep all intermediate files. may take lots of space!
examples:
analyze_simulation.pl -verbose -zrange=0:10:1 run1_cafl.i2i > & sim.out
xmgrace run1_cafl.xy
matlab: data = readxmgr('run1_cafl.xy'); acurves(data);
analyze_simulation.pl -verbose run1_cafl.i2i run1_ca.i2i > & sim.out
calc_diffusion3.pl
This program reads in a 2D diffusion tracks and calculates a D for each file (track_\d+.out)
by fitting a line to the MeanSquaredDisplacement vs time:
MSD(t) = 4Dt (it would be MSD(t) = 4Dt + 2*sigma^2 if there is noise, see ref below).
so the slope of the line divided by 4 should yield D (even in the case with noise).
note: Lag intervals overlap one another unless -no_overlap is specified.
meanD, varD, and standard error of the meanD is printed to stderr.
note: if -no_overlap is not specified, the standard error of the mean (seD) may not be correct
since the points aren't independent.
calc_diffusion3.pl [options]
-minlag=# if not specified, minlag = 1
-maxlag=# if not specified, maxlag = 1/2 the total time in the first track file analyzed
-duration=# rather than analyze each track in its entirety, split it into (non-overlapping)
sections of the specified duration (number of time points).
-files=file1:...:filen list of filenames to analyze (instead of all track_#.out files)
-filerange=#:# only look at files track_#1.out to track_#2.out (inclusive)
-logfit also do the D calculation (via a line fit) in the log-log space (ln (MSD(t) = ln(4D) + ln(t))
-no_overlap
-print_Ds print all the Ds calculated, one per line (tracknum D), to stdout.
-print_wDs print all the weighted Ds calculated, one per line (tracknum D), to stdout.
-print_graph prints to stdout MSD vs time
-notracknum the filename being read does not have tracknumber in it, just autonumber them.
-dummy just print some info to stderr
-verbose which file is being analyzed printed to stderr
example:
calc_diffusion3.pl -print_graph files=track_1.xy | cull -c -s -1 -2 |xmgrs
# take a 1,000 pt track and analyze it as 10 separate 100 length tracks to see what those Ds look like:
calc_diffusion3.pl -print_Ds -print_wDs -files=track_1.xy
calc_diffusion3.pl -duration=100 -print_Ds -print_wDs -files=track_1.xy
see also:
calc_diffusion.pl same program but only operates on one file.
calc_diffusion2.pl same program but without the -duration option
SingleParticleTracking_DiffusionCoefficients_Saxton.pdf p. 1746 for the algorithm
SubdiffusionOfSingleParticleTracks.pdf - p. 4 gives the formula if there is noise in the estimate.
compare_volume_measurements.pl
This program combines data produced by cross_sections_of_voronoi (-vfile) with
an analysis of the same image within fiji (via the stereology_lml macro, -sfile).
Results printed (on each line there will be sfile data then vfile data) to stdout.
compare_volume_measurements.pl [options] -vfile=filename1.txt -sfile=filename2.txt
options:
-print_volumes rather than print entire combined lines, just print volume info in columns.
the columns are:
region: the region id, this is the same as the pixel value.
sarea: the cross-sectional region area as reported by the fiji stereology_lml.txt macro.
svolume: the volume estimated by stereology_lml.txt
swvolume: the weighted volume estimated by stereology_lml.txt (not really a true volume).
true_volume: the true volume of the region (as created by cross_sections_of_voronoi).
area: area as reported by cross_sections_of_voronoi
volume_from_area another way to estimate volume from area, w/out stereology: ((1/.68)*area)^(3/2)
-vprefix=# foreach field printed out, change its name by adding the specified prefix
-sprefix=# foreach field printed out, change its name by adding the specified prefix
this may be used so that field names become unique (otherwise, eg, there
will be two "area" fields and two "volume" fields on each line).
-verbose stuff to stderr
examples:
cross_sections_of_voronoi -P voronoi.i2i > filename1.txt
Fiji
Edit->options->conversions - make sure "scale when converting" is NOT checked.
Plugins-> open i2i -> voronoi.i2i, then Image->type->32, Image->type->16, to get rid of the table lookup
Plugins->macros->install... install stereology_lml.txt
Plugins->macros->stereology_lml.txt -> voronoi_fiji.txt
compare_volume_measurements.pl -vfile=filename1.txt -sfile=voronoi_fiji.txt
# look at volume estimated via stereology (x axis) vs. true volume (y axis):
compare_volume_measurements.pl -print_volumes -vfile=filename1.txt -sfile=voronoi_fiji.txt | cull -c -s -3 -5 | xmgrace -free -pipe
# look at volume estimated from area (x axis) vs. true volume (y axis):
compare_volume_measurements.pl -print_volumes -vfile=filename1.txt -sfile=voronoi_fiji.txt | cull -c -s -7 -5 | xmgrace -free -pipe
# compare true volume (x axis) to both the stereology estimated volume and the volume from area measurements (y axis):
compare_volume_measurements.pl -print_volumes -vfile=filename1.txt -sfile=voronoi_fiji.txt | cull -c -s -5 -3 -7 > tmp.nxy
xmgrace -nxy tmp.nxy -free
see also:
/home/lml/krypton/Corvera/Raz/stereology/README, run_voronoi.pl
cross_sections_of_voronoi
/home/lml/krypton/packages/fiji/Fiji.app/macros/stereology_lml.txt
filter_beads.pl
Reads in a file produced by makebeads2 (bead.info) and one produced by print_pixel_coords (mask.info).
A bead's info is kept only if at least one of its voxels is under the mask.
Kept beads are printed to stdout with their original values (bead id, position, etc.).
Thus, if you actually mask the bead image (using maski2i) which was produced by makebeads2, this will
keep the associated bead info for those beads which have at least one pixel left in the image after masking.
Note: this can only be run on data produced by versions of makebeads2 newer than 2/12/09.
filter_beads.pl bead.info mask.info > filtered_bead.info
options:
-keepcenter=# : rather than keep a bead if any of its voxels is under the mask, only keep the bead
if its center is under the mask. # is the scale factor to convert from the original
resolution the bead was created at (-S option to makebeads2) and the resolution of the mask.
For example: makebeads2 -S 0.005 0.005 0.005 ... > beads.info
If you then use a mask at .1 um you would specify -keepcenter=20
-verbose
Example:
makebeads2 -v -d 1280 1280 60 -S 0.005 0.005 0.005 -n 300 -i 1 beads300.i2i > bead.info
maskimage beads300.i2i mask.i2i beads_masked.i2i
# now want to filter bead.info to leave only beads at least partially under mask.i2i (i.e., beads with at least one pixel
# left in beads_masked.i2i)
print_pixel_coords -2 mask.i2i > mask.info
filter_beads.pl bead.info mask.info > filtered_bead.info
# now filtered_bead.info "matches" what is left in beads_masked.i2i
An example where resolution is changed:
# voxel coords printed to bead.info by makebeads2 will reflect the new (chres'ed) coords appropriately:
makebeads2 -v -d 1280 1280 60 -S 0.005 0.005 0.005 -c 0.025 0.025 0.01 -n 300 -i 1 beads.i2i > bead.info
# now actually change the res of the bead image to match:
chres -S 5 5 2 beads.i2i beads_ch.i2i
# apply the mask to the lower res image. mask must already be at that same resolution:
maskimage beads_ch.i2i mask_ch.i2i beads_ch_masked.i2i
# get out coords of all pixels in this low res mask:
print_pixel_coords -2 mask_ch.i2i > mask_ch.info
# now just keep the bead info for beads which are under the mask at this low resolution:
filter_beads.pl bead.info mask_ch.info > filtered_bead.info
# now filtered_bead.info "matches" what is left in beads_ch_masked.i2i (I hope!)
See also: makebeads2, print_pixel_coords
graph_xpp2.pl
Reads specified columns of data from an xpp output file. Sends output stdout. note: time is always the first variable output (i.e., the first column). usage: graph_xpp2.pl [options] output.dat > name1_name2.dat options: -allstates: extract all columns of data having to do with channel state. This is all the basic data. Other data is derived from this. -openstates: all open states. cannot be specified with -closedstates. -nopen: sum of all the open states. cannot be used with -closedstates. -closedstates: all closed states. cannot be specified with -openstates. -verbose example: xpp -silent bk.ode -> output.dat graph_xpp.pl -vars=time:name1:name5:name7 output.dat > xpp_subset.out xmgrace -nxy xpp_subset.out see also: graph_xpp.pl
random_path.pl
Generate a random path (ie, diffusion). usage: random_path.pl [options] > track_1.out options: -D=# diffusion coefficient, default = 1 -length=# pathlength (number of time points). default = 1000 -xy only print x,y rather than t,x,y -method=oversample|gaussian oversample takes dt size substeps to calculate each 1 time unit step. gaussian just samples into an equivalent Gaussian distribution once for each 1 time unit step. default: oversample. gaussian option not tested yet... -dt=# substep time interval, default = .001 (only used if -method=oversample). -seed=# so random numbers are the same from run to run (only if -method=oversample) note: using the default random number generator which is probably not great. see also: run_random_path.pl calls random_path.pl to generate lots of paths. calc_diffusion3.pl can analyze these paths ~/krypton/diffusion/mytracks/
readbeads.pl
Reads in file produced by makebeads to check to see if sizes and amplitudes are Gaussian and Poission (-G and -P options). Produces _tmpsize and _tmpamp. You may want to use filterbeads.pl prior to using readbeads.pl. You can read these into xmgrace and then create histograms if so desired. readbeads.pl makebeads.out -skipzero skip values which are zero -outer put outer diameter (rather than "size") into _tmpsize. If makebeads used the -g option, then size always equals 1. outer should be the gaussian mean + 1/2 shell width. -rpts=# also prints out _coords.rpts file of coordinates of center of beads. This can be read in by addlines. Can also be read by pt_distances. # is a scale factor to divide by to get the new coordinates (so you can compare these to a lower resolution image). Set # to 1 if you don't want to scale the values. Note: coords are truncated an integer value (new = integer(old/# + 1)). skipzero command doesn't affect this option. note: if more than 1 point has the same new coordinates, then the w value for each duplicate point is 1 higher than the previous value (e..g, if -w=-10, then the first (5,6,3) has a w = -10, next has a value -9, next -8, etc. note: this also causes coords to be converted from 0-indexed (makebeads2 output) to 1-indexed (play assumes this for rpts files). -dontrescalez: when using -rpts=#, don't rescale the z coord -twod ignore z dimension when printing rpts file (still must specify -rpts option). -w=# in rpts file, set w to specified number, default = -1000 -amp use the amplitude of the bead for the w value in rpts file output See also: filterbeads.pl, makebeads2, objs2dist121.pl, print_pixel_coords, filter_beads.pl
run_random_path.pl
Generate lots of random paths files (ie, diffusion) by calling random_path.pl multiple times.. usage: run_random_path.pl [options] options: -seed=# so random numbers are the same from run to run -D=# diffusion coefficient, default = 1 -length=# pathlength (number of time points). default = 1000 -xy only print x,y rather than t,x,y -numpaths=# default = 10 paths -rootname=string default: track_ (so files will be called track_#.out) -verbose stuff to stderr note: using the default random number generator which is probably not great. see also: random_path.pl calc_diffusion3.pl
run_voronoi2.pl
This script calls cross_sections_of_voronoi many times, and produces an image and output text file from each run. run_voronoi2.pl [options] options: -num=# number of times to run. default = 10 -D=# -D option to cross_sections_of_voronoi, average diameter, default = 150 -dummy don't do anything, just echo to stderr what would be done
voronoi_vs_fiji.pl
compare the true volumes of voronoi regions (as reported by cross_sections_of_voronoi, e.g.
voronoi_D170_0001_g0_G250_volume.txt) to those calculated
within fiji using stereology_lml.txt (e.g., voronoi_D170_0001.txt)
Graphs volume (x) vs predicted volume (y) (or prints to stdout if -stdout is specified)
note: if looking at weighted volumes (eg, in weighted/ ), the individuals stereologic volumes
really are meaningless, and you should really just pull out the wmean values from each run.
usage:
voronoi_vs_fiji.pl [options] *volume.txt
options:
-vdir=/path/to/directory location of directory with the output from cross_sections_of_voronoi
(eg. voronoi_D170_0001.txt) default is the current directory
-debug info about all fields on each line printed to stderr
-stdout everything to stdout
-print_avg for each file, just graph mean and std error of the mean (gotten by combining all the data for each file, so
may include multiple stereological runs) rather than all data pts
note: this will average together the multiple fiji stereology runs which are often in the same *volume.txt file.
I think that is ok (but may make the sem look better than it would be from just one run).
-use_wmean just use the mean or wmean stored in the *volume.txt file and compare these values to ??? ... not done yet
examples:
cd /storage/big1/lml/Raz/voronoi
run_voronoi2.pl -D=170 -num=20 # calls cross_sections_of_voronoi
run_voronoi2.pl -D=220 -num=20 # calls cross_sections_of_voronoi
Fiji # then run run_stereolgy_voronoi.ijm
cd results/
voronoi_vs_fiji.pl -vdir=.. *D170*volume.txt # -> voronoi_vs_fij_D170.xmgr
voronoi_vs_fiji.pl -vdir=.. *D220*volume.txt # -> voronoi_vs_fij_D220.xmgr
see also:
/storage/big1/lml/Raz/voronoi/ - this is where I've done what is shown in the examples above
/storage/big1/lml/Raz/voronoi/README, true_vs_predicted.xmgr
/home/lml/krypton/Corvera/Raz/stereology/README, run_voronoi.pl, run_voronoi2.pl, compare_volume_measurements.pl, plot_results.pl
cross_sections_of_voronoi
/home/lml/krypton/packages/fiji/Fiji.app/macros/stereology_lml.txt
/home/lml/krypton/packages/fiji/run_stereology.txt
analyze_cross_sections_of_voronoi.pl
ball
This program creates an elliptical shell of brightness the specified brightness (-i option)
It is used to create test images for Fischer/hmc.
Usage: ball [options] ball.i2i
options:
-d xdim ydim zdim: dimensions of image, default = (50,100,50)
-r x y z: x,y, and z radius in voxels, default = (20.000000,20.000000,20.000000)
-s samplerate: number of subsamples in x and z directions per pixel, default = 1
-i intensity: intensity of the ball's shell, default = 100.000000
-b intensity: intensity background voxels, default = 0.000000
-f image: read in the specified image, add the ball image to that
(just replaces voxels along the ball's shell with new values).
-t thickness: thickness of shell, default = 1.000000
Note: you can call this multiple times to get a shell within a shell
code in /home/lml/krypton/facil/ball.c
calc_ca3
#/home/lml/krypton/bin/calc_ca3
Reads in 4 floating point images (or 4 short int images) as produced by simulation, calculates
the difference image deltaca.i2i (as a floating pt image) between the observed free calcium and what would be
expected in equilibrium conditions, i.e., [ca]-[predicted_ca], where [predicted_ca]
is kd*[CaFl]/[Fl]. Output concentration in nM.
All input concentrations should be in nM
Each z slice is analyzed separately (i.e., z is assumed to be time).
Usage: calc_ca3 [options] ca.i2i fl.i2i cafl.i2i cafixed.i2i deltaca.i2i
options:
-k #: kd in uM, default = 1.125000
-h: print cumulative histogram for each z slice.
options for printing graphs of free calcium (not concentration) vs. distance from ca entry location:
the units for the graph are nM*um^3, but 1nM = .6ions/um^3 , so multiply be .6 to get ions
(and 1pA = 3,000 ions/msec if you want to convert to pA-msec).
-c: print delta calcium (delta from first slice) distance graph for each z slice.
-n: normalize delta calcium distance graph for each z slice (so sum of amplitudes = 1),
(otherwise when graph spacing is changed, -g option, the height of the graph will change).
-a: print absolute calcium, not delta calcium.
when this is applied to a uniform image it should produce a 4pi*r*r distribution.
options for printing graphs of total calcium, i.e. ca+cafl+cafixed (not concentration):
-C: print delta total calcium distance graph for each z slice.
-N: normalize delta total calcium distance graph for each z slice (so sum of amplitudes = 1).
-A: print absolute total calcium, not delta total calcium
-p # #: pixel size in microns in the radial (x) and lengthwise (y) directions, default = 0.10 and 0.10
-P # # #: pixel size in microns in x,y,z directions (rectangular not cylindrical coords), default = ( 0.10, 0.10, 0.10)
-r: data is rectangular, not cylindrical coords (-P sets this too)
-e x y: 0-indexed coords of calcium entry location (default: x= 0, y = ydim/2)
-g #: graph spacing for -c,-C,-n,-N,-a,-A options. defauls is pixel size.
-s #: subsample each voxel this number of times in the x (radial) and y (longitudinal) directions,
default = 3. Subsampling only applies to histogram and graph generation, not image generation.
-z low hi dz: only look at z slices low to hi (0-indexed, inclusive), stepping by dz.
The zdim of deltaca.i2i will not change, but skipped slices will be 0.
see also:
float2int, in matlab: readxmgr, acurves (see ~lml/krypton/JSinger for an example)
source code in ~lml/krypton/Walsh directory.
cellsim
This program simulates Ca and Na in a cylindrical cell with
cylindrical symmetry.
Note: It is recommended that the stderr output be examined for
lines with the words: warning, Warning, error, or Error on them.
Usage
cellsim [options] >& output_datafile
Options controlling the duration of the simulation:
-time #: total time to simulate (in sec), default = 0.000000
-time_rate #: slow down time, ie take smaller dt steps during simulation.
this may be necessary if you are creating a large ca gradient,
for example, with -ca_ring or -ca_src options (.5 means 1/2 the dt).
The number of iterations needed for convergence is
roughly: I= (T*diff)/(dr*dr), where T is time, dr is
annulus thickness and diff is diffusion constant.
This ignores ion influx,etc. If you set diffusion rates very
small, the iter # calculated will be too small if other stuff
is happening, use -time_rate to compensate.
Options controlling the geometry of the model:
-radius #: radius of the cell in microns, default = 6.000000
-rings #: radius of the cell in rings, default = 60
Ring thickness = cell radius/ # of rings.
-length #: length of the cell in disks, default = 300
Each disk will be the same thickness as a ring.
-bc Rmax Lmin Lmax: boundary conditions at maximum radius and minimum and
maximum length coordinate (i.e., at the cell ends). These must
be the letters I (for infinite) or S (for solid, i.e. no ions go
through the membrane). The I option is not always accurate. default: S
-mask image.i2i: only voxels in image.i2i > 0 are part of the cell. Use this
to specify an arbitrary shape for the cell.
Not extensively tested.
-spacing file: file specifies the spacing along each axis (i.e, size of each voxel).
spacing is in nm, one entry per voxel, comment lines ignored (#).
The cell direction (X,Y, or Z) must be the first letter on the line.
Other characters on the line will be ignored.
The number of entries must be < 2000 each direction.
Do not specify -rings,-radius,and -length if you use this option.
NOTE: the simulation time step goes as (length)^2, where length is
the length of the shortest side of all the voxels specified.
Any number of entries per line. First R, then D (length). Example:
# R direction (10 pixels in this direction, high res at low R)
X
25 50 75 100 200 300 400 500 600 1000
Y (lengthwise) direction (9 pixels in D, very high res in the middle)
100 200 300 400 500 540 550 560 1000
Use cyl2cart with -spacing option to convert output images to
a uniform spacing. (I haven't added that option yet)
Not extensively tested.
Options specifying substances (ions, buffers, compounds) present in the model (10 max):
-ion name free_concentration D valence:
concentration in mMolar, D in cm^2/sec, valence in absolute value.
valence is only used if -ion_src is also specified.
concentration is the initial free concentration.
-buffer name total_concentration D:
concentration in mMolar, D in cm^2/sec.
concentration is the initial total concentration (sum of its free and all bound forms).
-kin bufname ionname boundname onrate offrate D:
kinetics for interaction between bufname and ionname. Boundname is the
name to give the bound pair. bufname and ionname must have been
specified (with -ion -buffer or -kin options) prior to this option.
onrate:in 1/(mMolar*seconds), offrate: in 1/seconds, D: in cm^2/sec
-equil_error relerr iter: maximum relative equilibrium error allowed during the
initial calculation of concentrations from the user specified
free ion concentrations and total buffer concentrations.
rel_error = ABS((true_kd - calculated_kd)/true_kd). Default = 1e-05
This should be small enough unless you have huge buffer
concentrations (>100 mM?). iter (an int) is maximum number of iterations
to allow during equilibration, max =1 billion, default = 10000000
Options controlling changing conditions:
-ion_src name current dur lowdisk highdisk lowring highring:
have a current source of the specified ion coming in through the specified rings
and numdisk (rings and disks are 0-indexed indices, inclusive).
Current is in pA for the specified duration (sec).
The current is uniformly divided within the entire specified input volume.
All -buffer and -ion options must be specified prior to this.
See comments by the -time_rate option.
-ion_src_file name imagename current duration:
Similar to -ion_src, but the specified floating point image is used to
scale the input current at each voxel. The total current coming into the
cell is adjusted so that after this per pixel rescaling, it will = "current".
-ion_current_trace with op = S, will modify this.
Typically do: int2float -M 1 psf.i2i imagename prior to this.
-ion_current_trace name filename dt op threshold: modifies current specified with
the -ion_src or -ion_src_file options.
Times which the channel is open (i.e., current flows) is specified in a file.
The file is a list of numbers (current or scale), 1 per line, at dt (in sec) intervals.
If op = T, currents greater than the specified threshold means channel open, otherwise closed.
If op = S, the current as specified with -ion_src is scaled by the number in the list.
"threshold" is required but ignored when op = S. Not extensively tested.
-zap_sub name time:
At the specified time (in seconds) set the named substance concentration
to 0. This does not prevent it from being produced from other reactions.
This is useful if an injected "ion" was photons (e.g., photon+caged -> uncaged)
and the laser is turned off at the specified time.
-ion_pump name max_rate n k lowdisk highdisk lowring highring:
have a pump of the specified ion remove ions in the specified rings
and numdisks (rings and disks are 0-indexed indices, inclusive).
max_rate * [ion]^n / (k^n + [ion]^n), [ion] and k in mM. max_rate in mM/sec.
max rates are typically given per unit area (8E-7 umoles/(cm^2 sec) for serca pump).
If we divide these published numbers by the ring width (typically 100 nm) we can get
a rough rate per unit volume. A typical max_rate might be .1 mM/sec (seems high,
Polito et al. biophysj, May, 2006 give a rate in terms of []/volume of ~ .001 mM/sec
- but they also have a k on the low end, .0001mM, which would compensate some.
Shuai & Jung, PNAS, Jan. 21,2003, p. 507 have a max rate of .5 (uM?)/sec.)
Typical values for k might be around .0001-.0005 mM. n=2 for serca pump (must be integer)
All -buffer and -ion options must be specified prior to this.
-ion_leak name rate compartment_conc lowdisk highdisk lowring highring:
have a leak of the specified ion introduce ions into the specified rings
and numdisks (rings and disks are 0-indexed indices, inclusive).
rate * ([compartment_conc] - [ion]), [] in mM. rate in 1/sec.
Leak rates are usually set to exactly balance the pump at initial ion concentrations.
compartment_conc for [ca] in the SR might be around 2 mM.
If max_rate = .001mM/sec, k = .0005mM, compart_conc = 2mM, and resting [ca] = 100nM,
rate should = .00001923/sec to balance the pump with the leak.
note: Leaks INTO the compartment (when [ion]_in_cyto>compartment_conc) are not allowed.
All -buffer and -ion options must be specified prior to this.
Options related to output:
-save name filename type scale dt:
Save images of the specified substance (name) to specified file. By default
substances are saved in mM. This will be multiplied by the scale
specified here. dt (in sec) is how often to save images. Type is I
(short int, our normal format) or F (floating pt,2x space, more accurate).
The first image saved will be prior to any ion current. The last image
will always be the end of the simulation. "name" should be as
specified with the -ion, -buffer or -kin option.
If the image file already exists, new images will postpended to it.
-save all tag type scale dt:
Equivalent to -save for all ions and buffers. Uses default file names,
but adds the tag on the end. ions in nM, all else in uM, then times scale.
-save_flow name rfilename lfilename scale dt:
Save flow images of the specified substance (name) to specified files.
rfile is radial flow, lfile is lengthwise flow. Images saved in floating pt.
Flow is in pA (1 pA = 3 molecules/usec) times scale.
dt (in sec) is how often to save images.
-save_flow all ltag rtag scale dt:
Equivalent to -save_flow for all ions and buffers. Uses default file names,
but adds the tag on the end. Output same scale as above.
NOTE: flow for EACH substance takes 8*(number of voxels) of space.
-debug #: set debug level to #. May substantially slow down the simulation.
1: extra print statements at init. 2: more testing while running. 3:
-debug_subvolume #: Normally the total mmoles (in the entire cell) of each saved substance is
printed to stderr. Setting this option will also cause the printing of the
total over a subvolume. Subvolume is all y >= # (0-indexed).
Example: if a cell has patch region at y=[1,10], setting # to the neck (11) of the
patch will let you see how much actually gets out into the main part of the cell.
Options related to MPI (multiprocessor stuff):
invoke on the wolves as, for example, mpirun -np 3 -machinefile file_of_machine_names cellsim ...
-print_proc #: only print output from the specified process (0 indexed).
Some output (eg, Total mmoles of ions, etc) are only valid for process 0.
-overlap #: overlap neighboring regions by the specified number of voxels (default = 50)
This will also be the number of iterations between necessary synchronization.
-debug_mpi #: print MPI info. Execute MPI dummy stubs if MPI NOT present (eg, under Windows,SGI).
This version of the executable does NOT use MPI.
-debug_mpi_proc rank numproc: set dummy rank and number of processes. use only if MPI NOT present!
EXAMPLES
./cellsim -time .0010 -time_rate .5 -length 200 -rings 40 -radius 1 \
-ion Ca 100E-6 2.5E-6 2 -buffer Fluo3 .050 2.5E-7 -buffer Buf .230 1E-14 \
-kin Fluo3 Ca CaFl 8E4 90 2.5E-7 -kin Buf Ca CaBuf 1E5 100 1E-14 \
-ion_src Ca 1 .00105 99 102 0 3 \
-save all _run1 I 1 .0001 >& cellsim_run1.out
Simulate for 100msec saving images every msec. 1 pA comes in the the pattern as specified
by psf_float.i2i for 10 msec. The current is then modulated by values in current.txt which
are one value per line, dt of 1 msec between them. Cell shape in mask.i2i. Voxel sizes in
spacing.txt
./cellsim -time .100 -time_rate .5 -spacing spacing.txt -mask mask.i2i \
-ion Ca 100E-6 2.5E-6 2 -buffer Fluo3 .050 2.5E-7 -buffer Buf .230 1E-14 \
-kin Fluo3 Ca CaFl 8E4 90 2.5E-7 -kin Buf Ca CaBuf 1E5 100 1E-14 \
-ion_src_file Ca psf_float.i2i 1 .010 \
-ion_current_trace Ca current.txt .001 S 999 \
-save CaFl cafl.i2i I 1000 .001 -save Ca ca.i2i I 1E6 .001 \
-save_flow Ca ca_radial_float.i2i ca_long_float.i2i 1 .001 >& cellsim.out
SEE ALSO
~lml/vision/bin/float2int (convert a floating point image to standard SHORT INT format)
~lml/vision/bin/int2float (convert normal image to floating point)
~lml/vision/bin/playexact (to display a floating point image)
~lml/invitro/Kargacin/changeheader (change header of floating point image)
~lml/vision/bin/cyl2cart (convert from cylindrical to cartesian coords)
~lml/vision/bin/nadiffl7 (many more options, but only 1 ion and 2 buffers)
~lml/vision/bin/diffuse2 (simple Gaussian spot diffusion program)
~lml/invitro/Kargacin/iplot (3D graph of a 2D image, intensity -> height)
~lml/invitro/Kargacin/blur4D_dFoverF (blur with psf, calculate dFoverF)
~lml/invitro/Kargacin/diffusion_limited_binding (simulate diffusion limited binding)
~lml/invitro/Kargacin/spot_diffusion (simulate diffusion from a spot)
~lml/vision/facil/printvals (print image values, calculate dFoverF)
~lml/vision/JSinger/src/channel (steady state based upon Neher paper)
channel_current2
channel_current2: like channel_current, but looks at [ca] over time at each BK (not just max [ca]).
From that, it calculates the max BK current. Channel_current adds the max [ca] that each BK voxel sees
from each voxel in a Ryr cluster, to get the max [ca] that BK pixel sees. But the max [ca] could occur
at different times since the Ryr cluster has a spatial extent. Experimental.
Calculates the current which would be produced when each object in Ryr_image.i2i releases calcium which
activates nearby BK channels in BK_image.i2i. Produces a histogram of these output currents.
The input images should have each pixel in an object having the object id as its value. These images can be
produced with "countobjs -Q" or "objs_via_ascent"
note: a one pixel border around the image is ignored (added 12/8/09).
This may take about one-half hour to run.
Usage:
channel_current [options] -f filename Ryr_image.i2i BK_image.i2i > im1_im2.hist
Options:
-f filename resting: file containing [ca], in nM, as a function of distance and time. Right now this is
assumed to have 20 nm spacing and entries every msec. See
/home/lml/krypton/Zhuge/STIC/simulations/run32.ca as an example
"resting" is the resting [ca], in nM, for the cell. It will we subtracted from the other
[ca] values to produce a "delta" [ca] which gets scaled linearly by Ryr intensity.
The spark current used to produce this (see -C) should be roughly in the range expected
for Ryr_image.i2i, since all delta [ca] is linearly extrapolated.
All distances > largest distance in filename will have their [ca] set to the [ca]
at the largest distance in filename.
note: Only the [ca] values at y == 1 are used. Is this what should be used?
-u #: voltage (mV). BK conductance is a function of voltage. default = -80.000000
-U low delta hi: voltages from low to hi by delta. Max is 20 voltages
-r #: resolution (binsize) for output histogram in femtoAmps, default = 1000.000000
-R #: reveral potential of BK channel in mV, default = -80.000000
distances:
-w # # #: x,y, zweights for pixel distances (ie, pixel size), default = 1 for each.
-p #: pixel size in nm (i.e., what 1 means in -w option), default = 80.000000
-m # : maxdistance (in units used for -w, i.e., pixels) to look for the closest pixel, default = 10.000000
which pixels or clusters to include:
-t # #: image1 and image2 thresholds, pixels > # are above threshold, default = 0.
-e lowx lowy lowz hix hiy hiz: only look at voxels within this extent (subvolume), 1-indexed, inclusive.
-s minsize maxsize: only look at objects in Ryr_image.i2i within the size range (inclusive, in voxels)
max size allowed = 100000
-S minsize maxsize: only look at objects in BK_image.i2i within the size range (inclusive, in voxels)
-i miniod maxiod: only look at objects in Ryr_image.i2i within the iod range (inclusive. use -d also.)
-I miniod maxiod: only look at objects in BK_image.i2i within the iod range (inclusive. use -D also.)
scale factors:
-d Ryrimage1d.i2i #: the original intensity image for Ryr_image (since Ryr_image has pixel values as id numbers)
# is the number of Ryrimage1d.i2i intensity units required to produce spark current
used in the calcium simulation (which produced the -f filename data).
This will scale the [ca] produced by each Ryr voxel. Default = 30000.000000
-D BKimage2d.i2i #: the original intensity image for BK_image (since BK_image has pixel values as id numbers)
# is the BKimage2d.i2i intensity from one BK channel.
This will scale current produced. Default = 1000.000000
If you want the intensity of each Ryr or BK object to affect the ca release (for Ryr)
or number of channels (for BK) , you need to provide the original data by using the
-d and -D options. Otherwise all voxels in an object contribute equally (so large objects
have more receptors or channels than small ones).
-c #: BK single channel conductance in pS, default = 220.0
-C #: peak current (in pAmps) used in the simulation to produce -f filename, default = 1.000000
This is just used for output produced by -v to estimate spark current produced per Ryr cluster.
miscellaneous:
-v: verbose. produces list of each Ryr cluster and K current produced to stderr. Other stuff too.
To write this to a separate file do: (channel_current -v ... > stdout.channel) >& stderr.channel
-V: very verbose
Examples:
cellsim -spacing /home/lml/krypton/Zhuge/STIC/simulations/spacing2.txt .... : produces ca.i2i
printvalsL -spacing2.txt -R -t -i ca.i2i > ca.vals
kate ca.vals : delete fist comment line
objs_via_ascent -s 5 100 Ryr.i2i Ryrid.i2i > tmp
tail tmp : the mean IOD gives an idea of brightness needed to produce current used in cellsim, say 30000
objs_via_ascent -s 5 100 BK.i2i BKid.i2i > tmp
tail tmp : the mean IOD gives an idea of brightness of a BK cluster, eg mean= 10000, but assume that's 10 channels
channel_current -f ca.vals -w 1 1 3 -d Ryr.i2i 30000 -D BK.i2i 1000 Ryrid.i2i Bkid.i2i > ryr_bk.info
cull -s -c -1 -4 < ryr_bk.info |xmgrace -source stdin
See also:
closest_object closest_voxel objs_via_ascent /home/lml/krypton/Zhuge/BKandRyr/graph.pl,match.pl
graph_channel_current.pl, test_channel_current, /home/lml/krypton/Zhuge/STIC/simulations/run32
/home/lml/krypton/Zhuge/BKandRyr/analysis6/cluster_distances.pl, plot_mean_sd_curve.pl
/home/lml/krypton/Zhuge/xpp/ASM15.ode
Bugs:
Source code in ~krypton/lml/facil
# Popen is monotonic as it should be.
must provide -f cafilename
cluster
Drops random Ryr receptors into a world. Then calculates how many clusters and their sizes get produced.
Currently the "world" default size is (2um)^3. (see -wsize)
The origin of this world is in it's center (so it goes from -1um to +1 um in each direction by default).
A histogram of cluster sizes is printed to stdout.
Usage:
cluster [options]
Options:
distance options:
-eucliddist: calculate a Euclidean distance (same in x,y,z directions). This is the default.
(use -cdist option to set the nearby distance for this).
-absdist: rather than calculate a real distance, calculate max(ABS(dx),ABS(dy),ABS(dz)) - for speed
(use -cdist to set the nearby distance for this.)
-psfdist x y z: distance (in um) a pt can be in the x, y, and z directions and be considered nearby.
Does not use -cdist. To approximate a psf you might specify .2 .2 .6
-cdist #: how close (in um) two receptors must be to cluster, default = 0.500000
other options:
-2D: do a 2D simulation (x and y), not a 3D simulation
-cylinder inner outer: only place Ryr within a cylinder (e.g., plasma membrane) with specified
inner and outer radius (in um). The cylinder has its length the entire y dimension.
Only the parts of the cylinder which fit in the world (-wsize) are used.
-wsize x y z: the width of the world (in um) in the x, y, and z directions (default = 2.0,2.0,2.0)
-psize x y z: pixel width (in um) in the x, y, and z directions (default = 0.1,0.1,0.1)
-numryr #: number of Ryr receptors to drop, default = 1000
-mindist #: when placing Ryr receptors, keep this minimum distance (in um) between them.
-randstart #: start the random number generator with # as the seed. So runs can be reproduced.
-image name.i2i write out an image of the Ryr receptors. Can then use blur3d to create a realistic
microscope image, and countobjs to analyze it and compare to real data.
diagnostics:
-debug: turn on debugging
-verbose: be a bit more verbose
-h: print this help
-H: print this help
-help: print this help
Examples:
Source code in ~krypton/lml/Zhuge/Ryr_clusters
/home/lml/krypton/bin/cluster -h
diffusion_limited_binding
unknown option h Prints out distance (um) and concentration (particles/cm^3) at that distance a slab volume of initial uniform concentration (1 particle/cm^3)which has a ligand binding to a plane at x = 0. This is diffusion limited binding. See my notes of 10/20/98. Usage: diffusion_limited_binding [options] options: -D #: diffusion (cm^2/msec), default = 2.2E-09 -t t1 t2: start and stop time to simulate (msec), default = 0.000000 1000.000000 set both numbers equal if you only want one time point. -x x1 x2: start and stop distances to print (um), default = 0.000000 10.000000 set both number equal if you only want one position -L #: thickness of slab volume (um), default = 10.000000 -T #: number of samples in t, default = 10 -X #: number of samples in x, default = 1000 -h: print this help message. -F: print out time (msec) and flux (particles/cm^2-msec) at x = 0 (i.e., rate molecules binding to x=0 surface). I've made flux towards lower x positive (normally it would be negative). -B: print out time (msec) and total # of particles/cm^2 bound at x = 0 (i.e., brightness of surface). Integral of flux from t1 = 0 regardless of -t option setting. -s: don't print out concentration as a function of x (i.e, the entire slab)
ellipse
This program creates an elliptical cell, cell length is in the y direction. It is used to model a squashed cell, and see how different it looks after blurring than a cylindrical cell Usage: ellipse [options] ellipse.i2i options: -d xdim ydim zdim: dimensions of image, default = (50,100,50) -r xradius zradius: radius (in pixels) in the x and z directions, default (20.000000,20.000000) -s samplerate: number of subsamples in x and z directions per pixel, default = 1 -i intensity: intensity inside the ellipse, default = 100.000000
em
Creates (or reads in) a HMM and then generates random data from it
(each model is a poisson process, with the j_th model having a lambda = 10*j).
Then attempts to use an EM algorithm to estimate the original model.
This is a way for me to learn how to implement the EM algorithm.
Usage: em [options]
options:
-s #: number of states, default = 2. Must specify -t and -p after this.
-t # # # ... #: state transition probabilities. If there are S states (e.g., -s #)
order: T[1][1], T[1][2], ... T[1][S], T[2][1] ... T[S][S] if there are S states),
where T[i][j] is the probability of a transition from state i to state j.
-p # # # ... #: prior probabilities of states (S entries if there are S states)
-P #: which prior estimate to use for the pdf of the data (ie., estimate of Poisson process)
1 = uniform, 2 = delta, 3 = uniform+delta, 4 = peice of observed data histogram (default)
5 = true pdf (Poisson)
-n #: number of data points to simulate, default = 100
-m #: maximum number of iterations to perform, default = 20
-truetrans: use the true transition probabilities as the initial guess (default = uniform)
-trueprior: use the true prior probabilities as the initial prior guess (default = uniform)
-findstates: also find (and print) the estimate of the states at each time point.
-v: verbose. print more info to stderr.
-V: print some more info. Some printouts only occur if # data pts is < 1000
Source code in /home/lml/krypton/facil/em.cc
See also: ~/krypton/facil/transpose.pl to print out selected lines from the output file
~/krypton/matlab/netlab: has EM routines
~/krypton/matlab/bayes/BNT/HMM: has HMM and EM routines
/home/lml/krypton/bin/em -h
makebeads2
/home/lml/krypton/bin/makebeads2 randomly distributes beads within a volume. created to let Jack simulate distribution of glut4 labels vesicles in the TIRF field of a microscope. A bead has a constant value between its inner and outer diameters. This value falls to zero 3 standard deviations inside the inner diameter and 3 sd outside the outer diameter. Beads are not allowed to intersect each other. Writes info about beads created or moved to stdout (can be read back in with -b option). note: coords written out are zero indexed (not 1-indexed like addlines and play) in pixels. usage: makebeads2 [options] outimage.i2i options: -d x y z: image dimensions in pixels, default = (256,256,64) -D # #: bead inner and outer diameters in microns, default = 0.200000 and 0.200000 -R #: inner rise standard deviation (in microns), default = 0.012500 -F #: outer fall standard deviation (in microns), default = 0.012500 -S x y z: x, y, and z pixel dimensions (in microns), default = (0.040000,0.040000,0.010000) -c x y z: x, y, and z pixel dimensions for new image size. If you are planning on taking the image produced by makebeads2 and using chres to change pixel size, put the new pixel sizes here. This is used when calculating the intensity of the brightest pixel from each bead. This output info can be useful for signal to noise ratio tests. -X #: number of subpixels per pixels, default = 10 -p # val: set z plane # (0 indexed) to val (modelling diffuse fluorescence on the plasma membrane) If val is < 1 it is interpreted as a fraction of total light in the image. For example: val = .2 means set the z plane to the intensity necessary so that the total light in that plane is 2012f the total in the final image (i.e., after exponential if -e or -E) -e dist: apply an exponential scale factor exp(-z/dist) to each zslice; like a TIRF excitation intensity dist is the 1/e distance in microns (.1 is a good value) This exponential also get applied to z plane value set via "-p # val" if val >= 1. It does NOT apply to that if val < 1. -E amp2 dist2:apply a second an exponential scale factor amp2*exp(-z/dist2) to each zslice must also use -e option. Each intensity will be scaled by (amp1*exp(-z/dist)+amp2*exp(-z/dist2)). amp1 + amp2 = 1 and is automatically set when -E is used (amp1 = 1-amp2). options related to output: -i #: outimage.i2i should be scaled by # before writing it. Format is short int (our default format). -f #: outimage.i2i should be scaled by # before writing it. Format is float. -v: verbose. -A: don't create outimage.i2i (still need a dummy name on command line though). -B: don't even simulate voxels (just prints beads centers, amplitudes, sizes). Automatically sets -A. -u file.objs: create file, which has a list of bead objects created, in the same format countobjs produces. this lets utilities written to analyze files in that format to then be used (e.g., objs2dist.pl, objs2bb.pl). COM field is the bead center even if part of it is outside the image. Size field is in pixels (only counting what is within the image). IOD is the total bead light in the image after exponential weighting (if -e or -E) and scaling (-i, -f). AVG = IOD/size. MAX is the max pixel (after -e,-E,-i,-f) . Some file header info may be wrong. -U file.objs: like -u, but all z coords are replaced by 1, and coordinates are for the new resolution specified by -c option. MAX value and max coord come from a sum projection in z at the new resolution. (after -e,-E,-i,-f). -W: print some warning messages to stderr if appropriate. note: This will mess up the output in the file if you are running it as: makebeads2 -W .... >& file.out options controlling randomization (of a uniform random distribution unless otherwise noted): -s min max: mininum and maximum range of allowed bead sizes. This number scales the bead inner and outer diameters (does not change sd). default = 1.000000 and 1.000000 -G mean sd: make bead sizes (scales) a Gaussian variable with specified mean and sd (instead of -s). note: since beads can't intersect, if you place beads densely you will tend to get more small beads (since the program generates a new random bead if one doesn't fit), and hence the size distribution will no longer match the parameters specified here. -g mean sd shell: like -G, but also specify bead shell thickness in microns. mean doesn't scale the shell thickness. mean is the diameter to midway between the inner and outer diameters (so don't use -D option). -a min max: min and max amplitude (brightness) of bead, default = 1000.000000 and 1000.000000 a pixel which is entirely covered by the shell of the bead (between -D numbers) will have this value. So this is like specifying a fluorescent concentration. -P lambda scale: make amplitude a Poisson variable with the specified mean (lambda) value (instead of -a option). this value then gets multiplied by scale. Amplitudes of 0 get ignored (a new amplitude is calculated). note: this is still just the amplitude of a covered pixel, not of the entire bead unless -T used too. -T: The amplitude specified with -a or -P option is the total light in the bead, not the light of one fully covered pixel. Takes about double the time to run. note: if the amplitude isn't large compared to number of pixels in the bead's shell, then individual pixel intensities will be small and truncation may effect total intensity. -x min max: allowed position of beads, in pixels, zero indexed, inclusive. The entire bead must fit in this region unless -o option also specified. -y min max: allowed position of beads, in pixels, zero indexed (default = anywhere within image) -z min max: allowed position of beads, in pixels, zero indexed (default = anywhere within image) -o: allow bead centers to fall outside the image volume (except in low z direction, i.e. coverslip). This is a more accurate way to represent a small view of a large cell. This option is ignored if -b option used. -O: like -o, but beads in low z direction outside the volume reflect back inside the volume (i.e. get moved back inside volume instead of thrown away). note: if bead density is too high, reflecting a bead inside may cause it to intersect with a previous bead, in which case the reflecting bead will be thrown away and a new randomly placed bead created. -w #: use # as the seed for random number generator, so can reproduce exact runs. must be negative. -b file xyspeed zspeed: read initial bead params (e.g., size, position) from file, just add random position to this. the random position uses the speeds (in microns) as the max the vesicle can move in each direction in this time period (i.e., dx = ran(0 to 1)*xyspeed, etc). Set xyspeed and zspeed = 0 if you don't want to move the beads. -j #: long int starting seed for random number generator. specify the same number (not 0) each time you run this program if you want to exactly repeat the random numbers generated. The default is to use a different starting seed each time based upon the process id when you run the program. -n #: number of beads to make, default = 100, max = 10000 Examples: #create 200 beads, with a 50 nm (5 z plane) gap before the pm, which has an #intensity of 50. Apply a TIRF exponential excitation scale to it. makebeads2 -e .1 -z 5 63 -p 5 50 -n 200 beads1.i2i > beads1.info # apply microscope psf blur3d -P -N 256 256 128 beads1.i2i empiricalpsf_seg_40nm_seg.i2i beads1_blur3d.i2i # bin (averge) to 100/160 nm resolution reduceima -A -X 1 256 4 -Y 1 256 4 beads1_blur3d.i2i beads1_blur3d4x4.i2i # extract infocus plane segment -Z 16 16 1 beads1_blur3d4x4.i2i beads1_blur2d4x4.i2i # add ScratchNG camera noise noise -e 1.4 -n 8 beads1_blur2d4x4.i2i beads1_blur2d4x4N.i2i histima -size 10 -bins 100 beads1_blur2d4x4N.i2i > beads1_blur2d4x4N.hist #creates beads, move them, look at the results makebeads2 -z 5 63 beads1.i2i > beads1.info #read those beads in and move them. but still must stay outside the first 50 nm. makebeads2 -z 5 63 -b beads1.info .05 .05 beads2.i2i > beads2.info concateima beads1.i2i beads2.i2i beads.i2i imsets beads.i2i 2 dave -tdim 2 -4 -I beads.i2i #just create scripts (very fast) with positions of beads, and move them around. makebeads -z 5 63 -n 200 -A dummy.i2i > beads1.info makebeads -b beads1.info .05 .05 -z 5 63 -A dummy.i2i > beads2.info makebeads -b beads2.info .05 .05 -z 5 63 -A dummy.i2i > beads3.info #now create the images makebeads -b beads1.info 0 0 beads1.i2i > beads1.info2 makebeads -b beads2.info 0 0 beads2.i2i > beads2.info2 makebeads -b beads3.info 0 0 beads3.i2i > beads3.info2 see also: makebead, readbeads.pl, blur3d, project, /storage/big1/sh/simul source code in ~lml/krypton/facil
oxygen
Calculates steady state partial oxygen pressure (P) as a function of distance from a blood vessel (r).
The blood vessel has a radius of R1 (5.0 um) and Oxygen at the vessel is Ps (52.0 mm Hg).
The default is to print partial Oxygen pressure (mm Hg) as a function of distance (um) as columns to stdout.
All models set P at R1 (and r<R1) to Ps. All models clamp P at R1 to be Ps for all time (vessels stay oxygenated).
The line source model (-m 1) implements equation 17 from KroghOxygenDiffusionModel1.pdf
Modeling pO2 Distributions in the Bone Marrow Hematopoietic Compartment.
I. Krogh's Model by D.C.Chow, L. A. Wenning, W. M. Miller, and E.T. Papoutsakis
Biophysical Journal, vol. 81, August 2001, pp. 675-684.
This models O2 assuming a constant oxygen consumption by the tissue and constant O2 (Ps) at the blood vessel.
They claim results don't change by more than a few percent using proportional (or other) O2 consumption models.
The value of dP/dr at R2 is forced to 0.
note: this is an implmentation of O2 from a line source (e.g., blood vessel), not a point source.
So it is not appropriate to convolve (in 3D) this with a 3D image of blood vessels.
But -p (or -m 2) will produce a point source image.
The point source model (-m 2) implements equation 14 from HeatConduction.pdf
Steady Heat Conduction in Layered Mediums: The Half-Space and Sphere, by Henry N. Pollack
J. of Geophysical Research, vol. 70(22), Nov. 15, 1965, pp. 5645-5648
This models O2 assuming a constant oxygen consumption by the tissue and constant O2 (Ps) at the blood vessel.
note: The value of dP/dr at R2 is forced to 0 unless -b # is specified.
note: Since O2 consumption is constant, it might not make sense to use this in a convolution, since it isn't
linear.
Model 3 is a point source model, but O2 consumption by the tissue is proportional to the O2 level in the tissue.
I derived it from Crank's equations 6.60 (infinite hollow sphere model) and 14.13 (see below).
It requires the specification of an initial O2 level in the tissue (P0, which is the same everywhere).
R2 is used only to determine how far out to plot the graph (and dP/dr at R2 is not used either).
See my paper notes or ~/krypton/Corvera/Olga/SteadyStatePointDiffusion.pdf
Not only is proportional O2 consumption more biological, but I think this should be linear and therefore
it makes more sense to use in a convolution with a (binarized?) blood vessel image.
note: because R2 really should vary in an image, depending upon the distance between nearby blood vessels
and because Ps is not just a scale factor (so, eg, brighter blood vessels in an image, if brightness is
proportional to Ps, do NOT just cause a scale factor in the P(r) values), it may NOT be appropriate
to take the image produced by this program and convolve it with an image of blood vessels to get
estimates of oxygenation of tissue. This comment may apply to all three models (definitely to -m 1)
Usage: oxygen -m # [options]
required:
-m # which model to use.
-m 1 :line source model with constant O2 consumption
-m 2: point source model with constant O2 consumption.
-m 3 :point source model with O2 consumption proportional to O2 concentration.
Use point source models if planning on convolving resulting image with a blood vessel image.
options for all models:
Although these options can be used in all models, different models have different sensitivities to them.
-R1 # vessel radius (um). Changes to this can change P a lot (if -m 1), and may require R2 to change (see -c).
default = 5.00
-R2 # max distance any tissue is from a vessel (um). This changes the solution a lot for some models
but for the -m 3 model it only affects how far out data is graphed, default = 2268.00
More precisely, for -m 1 and -m 2 it is the distance at which dP/dr = 0 (ie, a b.c. on the pde).
-Ps # partial pressure of Oxygen at the blood vessel (mm Hg). Default = 52.00
40 is typical of veins, 95 of arteries. This is held constant over time in all the models right now.
For -m 1, this will affect the O2 value at R1, but how P changes RELATIVE to P(at R1) (as r increases)
will not be affected (ie, it does NOT just scale everything).
model 1 options:
-c use with -R1 to automatically calculate R2 based on the specified R1 (since R2 affects the solution).
(this also sets -m 1 as a convenience)
model 2 options:
-b # changes the 2nd boundary condition to be P at R2 = # mm Hg
instead of dP/dr = 0 at R2 (this also sets -m 2 as a convenience)
model 3 options:
-P0 # the initial oxygenation level of the tissue (mm Hg). Default = 0.000000
This will change the solution a lot, since it is also the level that O2, in steady state, approaches as r->infinity.
Therefore, nonzero P0 may also make convolution with a blood vessel image inappropriate.
-P0 0 seems to produce the same results as -r. (-P0 also sets -m 3 as a convenience)
output graph options:
-n normalize output (doesn't affect -i image.i2i), ie, print r/R1 vs P/Ps rather than r vs. P
-r add a third reference column which is the "simple" diffusion solution (goes as 1/r)
This is for a point source in 3D (not a line source), and ignores O2 consumption.
graph as: oxygen_krogh -r | xmgr -nxy stdin
output image options:
-i image.i2i xdim ydim zdim: create an image of the values, to use with blur3d to find
O2 everywhere in an image as a function of distance from blood vessels.
If you are planning on convolving you should probably specify -m 3 .
-d x y z if -i is specified. pixel dimensions in um. default = 0.60 0.60 10.00
-s # if -i is specified. scale output intensities by #. Otherwise max is 52.0 mm Hg
-o xorig yorig zorig: location of the blood vessel in the image (zero indexed, floating point)
blur3d may care about this, depending upon options. default = (0.00,0.00,0.00)
(maybe I should change this to define a line in the image when -m 1 is specified?)
-p the output image should be for the simple 1/r point source (see -r explanation) rather
for the line source (-m 1) or point source with O2 consumption (-m 2 or -m 3).
other options:
-v verbose. stuff to stderr.
-V more verbose. stuff to stderr. debugging info.
-h print this help
Examples:
# create a binary image of blood vessels (maybe use BestPath3D_ellipse instead? Maybe thin3D afterwards?):
mask_image -m 2700 -r 1 G03_x_1_2_r4.i2i G03_x_1_2_r4.i2i G03_x_1_2_r4_binaryvessels.i2i
# create a psf of blood oxygenation:
oxygen -s 500 -i O2ptsource.i2i 50 50 21 -d 2.58 2.58 10 -o 25 25 10 -m 3 -r > O2ptsource.xy
blur3d -S .002 -Z -d -v G03_x_1_2_r4_binaryvessels.i2i O2ptsource.i2i G03_x_1_2_r4_O2.i2i
see also: /home/lml/krypton/Corvera/Olga/README, SteadyStatePointDiffusion.pdf (my notes)
Random Walks in Biology (New, expanded edition) by Berg p. 23 eq. 2.11;
The Mathematics of Diffusion (2nd Ed) by Crank p. 32 eq 3.5b; p. 89 eq 6.5; p. 102 eq 6.60; p. 330 eq 14.13
source code in: /home/lml/krypton/facil
error: A model must be specified, i.e., -m # must be specified.
sparkspread
Calculates the spread of spark event amplitudes given that two events occur at the same spark site and compares it to the spread of amplitudes given that two events occur at different spark sites. A spark site has a unique (x,y) position. Prints mean and stddev to stdout. Also prints out histograms and cumulative distributions. A valid datafile has either blank lines, comments (start with #) or a data line (spark event). A data line has 8 or 11 fields. 11 if a stoc is associated with the spark event A valid stoc (last 3 fields) must have a nonzero stoc amplitude (1st of 3 fields) See /usr/people/lml/vision/Rhonghua/Sparks/singlesitev2 for a valid file format. Usage: sparkspread [options] datafile options: -h: do NOT print histogram for intraspark data. -H: do NOT print histogram for interspark data. -c: do NOT print cumulative distribution for intraspark data. -C: do NOT print cumulative distribution for interspark data. -b #: number of bins for histogram, default = 20 -n: normalize the histograms so their maximum is 1 -k: also print ks statistic showing if two distributions are different.
total_ca
#/home/lml/krypton/bin/total_ca
Reads in a floating point image or short int image as produced by simulation (i.e., cylindrical coords), calculates
the total calcium change (i.e., [ca](r)*pi*r*r - t(0)) as a function of distance from the calcium entry site.
Output is printed to stdout.
All input concentrations should be in nM
Each z slice is analyzed separately (i.e., z is assumed to be time) unless image is rectangular and 3D (-R).
Usage: total_ca [options] ca.i2i
options for printing graphs of calcium (not concentration) vs. distance from ca entry location:
the units for the graph are nM*um^3, but 1nM = .6ions/um^3 , so multiply be .6 to get ions
(and 1pA = 3,000 ions/msec if you want to convert to pA-msec).
-n: normalize calcium distance graph for each z slice (so sum of amplitudes = 1),
(otherwise when graph spacing is changed, -g option, the height of the graph will change).
-a: print absolute calcium, not delta calcium (the default).
when this is applied to a uniform image it should produce a 4pi*r*r distribution.
The default, delta calcium, prints the delta (i.e., change) from slice 0 (regardless -z option)
(but delta calcium not calculated if 3D rectangular, i.e., -R specified).
-p # #: pixel size in microns in the radial (x) and lengthwise (y) directions,
default = 0.10 and 0.10
-P # # #: pixel size in microns in x,y,z directions (rectangular not cylindrical coords),
default = ( 0.10, 0.10, 0.10)
-r: data is rectangular, not cylindrical coords (-P sets this too)
(e.g., already passed through cyl2cart).
-R: data is 3D rectangular (not 2D+time), not cylindrical coords.
-e x y: 0-indexed coords of calcium entry location (default: x= 0, y = ydim/2)
-E x y x: 0-indexed coords of calcium entry location for 3D rectangular data
(i.e., z direction is not time). sets -R option.
-g #: graph spacing. default is the pixel size.
-s #: subsample each voxel this number of times in the x (radial) and y (longitudinal) directions,
(and z if 3D and not 2D+time) default = 3 (anything less tends to introduce errors).
-z low hi dz: only look at z slices low to hi (0-indexed, inclusive), stepping by dz.
see also:
printvals, float2int, in matlab: readxmgr, acurves (see ~lml/krypton/JSinger for an example)
source code in ~lml/krypton/Walsh directory.
vcspat-models
argc = 1
This program creates a volume image with objects randomly placed
within it. The mask image defines where an object is allowed to be placed.
The mask value can be changed with the -M option. Objects can either be individual
voxels (the default) or tiny images (-o option) or connected voxels in an image (-O).
Usage:
vcspat-models [options] mask.i2i out.i2i
Options:
-m # : model number (default 0).
0 - grid pattern
1 - random pattern
-M # : value of mask pixels (default 1).
-r # : number of random points to use in random pattern (default 25000)
-o name : image to use at point in pattern (default just turns on a pixel)
(can use up to 256 images - image will be randomly chosen)
-a : if objects overlap, add their pixel values instead of just replacing
Options related to objects:
-O thresh name.i2i: threshold an image to use for objects. Extracts as an object each
connected set of voxels above threshold. Places these objects randomly
into the output image (uses each object once). Use with the
-m 1 option. Do not use the -r option
-P name.i2i id.i2i: similar to -O option, extracts as an object each of the
the objects in name.i2i based upon their pixel values (object id) in id.i2i
(e.g., id.i2i was produced via objs_via_ascent). This option preserves the
distinction between objects in name.i2i which may be touching - since simple thresholding is not used.
If only an id.i2i is available (no name.i2i), you can specify -P id.i2i id.i2i
-v: replace objects (after fitting them into the image) with just the single voxel
at the brightest location in the object. Only use with -O and -e options.
-S # #: minimum and maximum object sizes to allow (use with -O option).
-e: do not allow (exclude) objects to be placed so that they overlap with each
other, go outside the image, or go outside the mask. The default is to allow
overlaps and to just zero out that part of an object which goes outside the
image volume or mask.
-n: number the objects in out.i2i (use with -O or -P options). Rather than all
the pixels in an object being their original values, all the pixels in object 1
will be labeled 1, all those in object 2 labeled 2, etc. If -e not specied and
objects overlap the first object placed (usually the largest) will prevail.
Can also be used with -v, then output will have the brightest voxel set to object id, others to 0.
The output of this can be analyzed by object_overlap program.
-N out2.i2i: create a second output file, number the objects in out2.i2i (like -n).
OtherOptions
-s # : seed for random number generator (default pid)
-x # : x increment for grid pattern (default 3)
-y # : y increment for grid pattern (default 3)
-z # : z increment for grid pattern (default 3)
-V: verbose
note: entire image name (with extension) is required.
note: a - in place of an image name means stdin or stdout.
Examples:
objs_via_ascent in1.i2i inid1.i2i
objs_via_ascent in2.i2i inid2.i2i
vcspat-models-new -m 1 -M 1 -P in1.i2i inid1.i2i -N outid1.i2i mask.i2i out1.i2i
vcspat-models-new -m 1 -M 1 -P in2.i2i inid2.i2i -N outid2.i2i mask.i2i out2.i2i
object_overlap outid1.i2i outid2.i2i > obj_overlap
overlapL out1.i2i out2.i2i > coloc
See also: krypton/Qiong/run1, vcspat, randompuncs, spatial_stats2, objs_via_ascent, countobjs
Source code in /storage/big1/lml/jac/was_invitro/Projects/cspat-volume
Image Analysis
Corecell.pl
Takes a 3d image and an outline and keeps all data pts within the specified distance
of the outline (removes the "core" of the cell).
Also produces a file named in_image_H.histo. See help for histogram2 about that.
If in_image2.i2i out_image2.i2i are specied, in_image2.i2i is also cored
(and histogram2 is applied to the pair in_image.i2i and in_image2.i2i when producing in_image_H.histo).
Written to let Ronghua only keep BK and Ryanodine label near the plasma membrane.
Note: touching voxels may have a distance of .5 voxels, and voxels at the same
position may have a distance of -.5 (??)
Note: pixels may be kept near the end of the cell, since they have close distance to the end
(rather than to the plasma membrane. Might -Z # # option to histogram2 fix this?).
Note: there might be problems if boundary.pts touches the edge of the image.
Corecell.pl [options] in_image.i2i boundary.pts out_image.i2i [in_image2.i2i out_image2.i2i]
options:
-interp interpolate missing z slices in boundary.pts before using it.
zslices must be in ascending order in boundary.pts.
contours must all be drawn in the same direction (i.e., all clockwise or all counter-clockwise).
The name of each object is changed to "nuke". A maximum of 1000 zslices is allowed.
-shrink=# shrink boundary.pts by this number of pixels before using it
-expand=# expand boundary.pts by this number of pixels before using it
-zrange=#:# only examine z slices (1-indexed in this range)
-dist=#: keep all voxels within this distance of the boundary, default = 2
-dx=# pixel size x size (default = 1. all that really matters is relative x,y, and z size)
-dy=# pixel size y size (although if you change this then that may affect -dist)
-dz=# pixel size z size
-dontcapends first and last outline slices aren't capped (removes -c from the call to histogram2).
typically you'd use this if the outlines are circular cross-sections of a cylindrical
cell (ie, z is along the length of the cell). If the outlines are along the length
of the cell (ie, cell is in the xy plane, as Jeff likes to do), then you wouldn't use this option.
-verbose print out extra info to stderr as it goes along
-vverbose very verbose
-keepfiles keep all intermediate files (all begin with _)
example:
#cell's length is in the xy plane, need to flip it to get cross sections to outline with planimeter3
transp -YZ im.i2i im_YZ.i2i
planimeter3 -I im_YZ.i2i -R im_YZ.pts
transp -YZ im2.i2i im2_YZ.i2i
#cell's original zspacing = 250 nm, but x and y are 80, so relative spacing is about 3:1, after transp
#it is the y direction that has the 250 nm spacing.
Corecell.pl -dy=3 -dist=3 -interp im_YZ.i2i im_YZ.pts im_YZ_near_pm.i2i im2_YZ.i2i im2_YZ_near_pm.i2i
dave -T 1 YZ -T 2 YZ -z 3 -I im_YZ_near_pm.i2i -I im2_YZ_near_pm.i2i
see also: shrink_wrap (to apply prior to Corecell.pl to get a better starting surface)
count_2d_objs2.pl
run countobjs on a 2D time series
creates temporary files _temp.i2i and _temp.stats in the current directory.
note: scale and black level forces each time point to be rescaled the same and
therefore to be thresholded the same.
file.stats will have data ordered as:
time number_of_objects mean_iod sd_iod mean_size sd_size
count_2d_objs2.pl [options] image.i2i file.stats
options:
-tstart=# first time point to examine, default = 1
-tstop=# last time point to examine, default = last z slice.
-min=# minimum size object to allow, default = 1
-max=# maximum size object to allow, default = 100000
-thresh=# threshold, default = 0
-scale=# scale, default = 1
-black=# black, default = 0
-allstats=filename print info about each object to _temp.stats (otherwise just summary stats from each t printed)
and rename it "filename"
-outimage=image.i2i see -o option in countobjs.
-Outimage=image.i2i see -O option in countobjs
-Qimage=image.i2i see -Q option in countobjs, note: id numbers will restart from 1 on each slice.
-verbose
-dummy
-noclean
distance2surface_overtime.pl
Finds the distance (in pixels) of a specified pt (over time) from the nearest pixel
greater than 0 in 3Dimage.i2i (or greater than -threshold). Alternatively, finds
the distance to the boundary surface specified with the -surface option.
The coords is as produced by track_pts (x y z otherstuff).
Written to let Andrea take a tracked gene in the nucleus and find its
distance to the nuclear envelope over time.
Note: touching voxels may have a distance of .5 voxels, and voxels at the same
position may have a distance of -.5 (??)
distance2surface.pl [options] 3Dimage.i2i specifiedpt.coords outfile.dist
options:
-surface=boundary.pts find distance to the surface defined by boundary.pts
rather than the distance to nearest >0 voxel 3Dimage.i2i
-shrink=# shrink boundary.pts by this number of pixels before using it
-expand=# expand boundary.pts by this number of pixels before using it
-zrange=#:# only examine z slices (1-indexed in this range)
-threshold=# threshold 3Dimage.i2i (voxels <= threshold set to 0, > threshold set to boundary).
-pixel=# pixel size (in x and y) in nm
-verbose print out extra into to stderr as it goes along
-vverbose very verbose
-keepfiles keep all intermediate files (all begin with _)
extract_area2.pl
Can't use 'defined(@array)' (Maybe you should just omit the defined()?) at /home/lml/krypton/bin/extract_area2.pl line 85.
findpaths_exo4.pl
this program takes output from exo4. similar to findpaths_exo3.pl, but parses exo4 output rather than exo3 output. usage: findpaths_exo4.pl file1.out options: which paths: -prestring="# debugpath 3 (102,156,-1):" pull out paths that begin with this. default is the null string. : null string paths should also be saved_paths later. : "# debugpath .*:" will pull out all the debugpaths (. matches any char and * means : number of them) -saved_paths : all the saved paths (within -zrange if that is also specifed) -debugpaths : all the debug paths. These options are mutually exclusive. -zrange=minz:maxz : only print tracks which start within the z range specified (1-indexed, inclusive) what to do with the paths: -saved_labels=filename : implies -saved_paths. saves labels for each path into the specified filename. This : file can be read into play so that labels are visible at a location. for rpts file: -rptsfile=track.rpts : create an rpts file (initially empty). use -rptstracks and -printfusion to put stuff in it. -rptstracks : put paths (tracks) into the rpts file specified with -rptsfile -printfusion=# : print box (of width #) around fusion events to rptsfile. must us -rptsfile option with this. : note: -debugpaths do not have valid fusion times. -pathcolor=# : instead of -1 -boxcolor=# : color of fusion box. instead of -2 for xmgr file: -trackxmgr=track.xmgr : create an xmgr file of the tracks produced by the program "exo". -truetime : rather than use z value as "time" calculate time based on the "burst protocol" : of 60 seconds acquisition at 1Hz followed by 3 seconds at 20HZ, repeatedly. -peak : rather than print avg (iod/size), print the peak pixel value (ie, value of maxima). -finalfit : rather than print avg, print the best fit curve (based on final fit parameters). -initfit : rather than print avg, print the init fit curve (based on initial fit parameters - for debugging). -distance : rather than print avg, print the distance from the first pt on the curve. -maxtracks=# : for -trackxmgr, only print out this many tracks. -tracks=start:skip : for -trackxmgr, only print tracks starting at track "start" (1-indexed) and : skip "skip" paths beforing printing another one (e.g., print every skip paths). : this lets you print a subset of paths which are still spread over the entire z range : to make visualization easier. skip = 0 means don't skip any. misc: -verbose -stationary_time=filename : print time each path is stationary prior to fusion to specified file example: exo4 -d 120 -o adp12273_ids.i2i -V -w 10 -b 2 adp12273_c1_z1201_1320_masked.i2i > & adp12273_c1_z1201_1320_masked.out play -min 0 adp12273_ids.i2i # maxima of each region should be blue findpaths_exo4.pl -trackxmgr=adp12273.xmgr -truetime adp12273_c1_z1201_1320_masked.out xmgr adp12273.xmgr #clicking on each set brings up its legend info which has its #starting coord (1-indexed I think) and value. findpaths_exo4.pl -rptsfile=adp12273.rpts -rptstracks -printfusion=4 adp12273_c1_z1201_1320_masked.out addlines adp12273_c1_z1201_1320_masked.i2i adp12273.rpts - | play -P -1 R -P -3 G - see also: ~lml/krypton/Huang/TIRF4/graph_exo3.csh
get_find_vesselness_results.pl
Can't use 'defined(@array)' (Maybe you should just omit the defined()?) at /home/lml/krypton/bin/get_find_vesselness_results.pl line 115.
mask_and_coloc.pl
Takes a threshold, x and y translation for vinculin image, a pts file, and two images, and creates a mask and then calculates colocalization. See README. mask_and_coloc.pl -x=# -y=# -thresh=# -pts=file.rpts -vin=vin.i2i -trip=trip.i2i options: -tx=# translate by a max of -tx when calculating other colocalization numbers, default = 25 -ty=# -step=# step size for other translations, default = 5 -noclean don't clean up (ie, don't remove) intermediate files created during this analysis -overlap=file just calculate overlap3, reusing the other files, put result into file.
mask_gedmat_images4.pl
mask_gedmat_images4.pl -check_intersect added.
determines when outlines intersect and sets the aerr (area error flag) in the
output file. eventually it will calculate areas a different way when that happens
rather than combining masked images (see ~/krypton/Corvera/Raz/README for 10/18/13). not done yet ...
Examines all the zip files (produced within fiji when outlining regions) in the current directory. These have
the boundary of the explant, day7 growth, and day11 growth.
note: this program assumes that all the data in the directory is from the same patient and the same tissue type.
It recalculates the area measures by only including that part of the day7 and day11 growth which is between the explant
center and going towards the center of the well; area from the explant center going away from the center of the
well (ie, growing towards the well wall) is excluded. By doing this, we hope to minimize the
effect of growth (as measured by area) being stopped by the well wall when the explant is near the well wall.
Output is written to stdout. The comments there explain the fields. area_mex is the old value, area_mex_border is the new.
This is done by finding all the *_Area.zip files in the current directory, pulling out the outlines in those files
and using that information to recalculate areas. It also needs to find the associated "combined" image file
(*_combined.i2i) to get image size and hence the position of the center of the well) and the fiji results file
(*_results.txt, so the numbers produced here can be easily compared with them).
note: The algorithm assumes that the area NEVER DECREASES with time, and that the explant region is always surrounded
by the day7 region which is surrounded by the day11 region. The latter assumption is sometimes invalid if
the day 11 images were shifted relative to day 7 images; but that is usually ok.
(if necessary, you can manually change the "bad_data" field in the output file to 1 and then use
-cond in gedmat_to_xy.pl to eliminate those stats from further analysis.)
It also assumes the smallest area in each zip file is the original explant area, the next largest area is
called day7 area and the largest area (if 3 are present) is assumed to be day 11 area.
Sometimes when growth is about 0, the outlines on, eg, day7, may have slightly less area than the original
explant, so the outlines might be mis-ordered then. [actually it may read the correct day off the name of
the i2i image now].
note: If only 2 areas (ie, two .roi files) are present in the zip file, they are assumed to be for
the explant and day7 (not day11).
note: This program assumes the center of the combined image (usually 4 well images are combined into one) is the
center of the well. see combine_images.pl. It also assumes the combined image name ends in combined.i2i
(see -combined).
note: some of the scripts I use to then analyze these results will multiply the area by 2, to get a "true" estimate
of growth; since the value produced by mask_gedmat_images3.pl will be (roughly) half of the real growth rates.
usage:
mask_gedmat_images3.pl [options]
options:
-tmpdir=fullpathname directory to use for temporary files which are created, eg: -tmpdir=/home/lml/krypton
-zip_file=file.zip
-zip_file=file.zip:file2.zip:...:filen.zip
only examine the one specified zip file, not all those in the directory
(If the second form is used, you can specify as many as you want.)
note: you may still need to specify -zip_ending so that the ending of this file can be stripped
off correctly (so the -combined_ending and -results_ending can then be added correctly).
-zip_ending=stuff e.g., -zipending=_area.zip . All files which end in this will be analyzed; default = _Area.zip
-skip=file.zip[:file2.zip...] list of zip files (without the full path) to skip.
-results_ending=stuff e.g., -results_ending=_results.txt . This is what the -zip_ending should be replaced by
to get the name of the associated fiji results file. note the _ should be included here
if it was included in -zip_ending.
-combined_ending=stuff e.g., -combined_ending=_combined_reduced_.i2i . default = combined.i2i.
This is any string which helps identify the last part of the combined image filename.
-combined_scaling=#:# e.g., -combined_scaling=2:2 if reduceima -bin 2 2 was applied to get _combined_reduced_.i2i.
-scale=#:# distance and area (eg, -scale=2:4 if reduceima -bin 2 2 was used) scaling. This will multiply
the area stored in the fiji file and the area calculated via my mask by the second number.
The first number will multiply the distance of the explant from the center of the image.
This option is just here to handle 001_BAP and 002_PRO datasets. They were analyzed in fiji
with the images reduced by a factor of 2 in x and y. So the fiji areas produced will be low
by 4x, as will my areas. This option compensates for that. See ~/Corvera/Raz/README for 10/11/13.
-check_intersect check if outlines intersect with the explant's outline. If so, set an error flag on stdout printout
and print info to stderr. eventually will just use area numbers rather than masking area in this situation.
that is not done yet ...
-save_day[=directory]
saves the final binary mask for each days growth (eg, day7 growth minus the explant and minus the part growing towards the wall)
into a 3d mask image (patientid_tissue_day#_mask.i2i). This image will be reduced down in xdim and ydim from the
original. patientid_tissue_day#_.play_labels will also be created (labels which well each z slice comes from).
This is mostly for quality control purposes.
In addition, patientid_tissue_day#.rpts will be created (similarly reduced down in size) (coords may be off by 1?).
View the file via: addlines maskim.i2i maskim.rpts - |play.FC14 -P -1 R -labels maskim.play_labels -
or via: play_results3.pl
patientid_tissue_day#_orig.i2i will be the original images (also reduced in xdim and ydim, and only one 2d slice
from each original 3d image).
note: older versions of play may need the comment line at the top of maskim.play_labels removed.
note: if you want images to be in another directory, specify a full pathname, eg -save_day=/home/lml/krypton (without last /).
note: any existing file with these names will be OVER-WRITTEN.
note: this option creates lots of temporary files (in tmpdir) of the form __*.i2i, any existing
files that match this will be DELETED.
see also: montage
-just_check just check whether each zip roi outline has the correct number of associated image files.
eg, if the zip file has 3 outlines (explant, firstday, secondday) there should be two i2i files with DAY# in their name.
prints to stderr info about those which don't have the correct number. does nothing else.
-verbose some extra info to stdout
-debug prints some tracing info to stderr
-print_fname print name of each zip file as it is being analyzed, to stderr.
-dlevel=# extra debugging. the bigger the number the more the printout. # is an integer starting at 1.
-dlevel=1 will print a trace of commands to execute to mimic (parts of) this perl script.
-noclean don't remove the temporary files which were created
the files are: _ones.i2i _explant.rpts _day11.rpts _day7.rpts _excluded_region.rpts
_explant.i2i _day11.i2i _day7.i2i _day7_noex.i2i _day11_noex.i2i _day7_noex_halfex.i2i _day11_noex_halfex.i2i
(if multiple zip files were analyzed, only the files from the last one will be left, since they get reused)
-dont_recalc only for diagnostics. does NOT restrict the analysis to just the growth away from the wall.
with this specified this entire program should just reproduce the numbers we already
have in *Results.txt since that uses the entire area. ie, this make the program a no-op.
see also:
/home/lml/krypton/Corvera/Raz/README
analyze_gedmat3.pl a script which calls mask_gedmat_images3.pl for all of Raz's data. Also versions 4,5,6, and 7.
play_results3.pl
raz_rates.pl
mask_gedmat_images.pl
mask_gedmat_images2.pl
/storage/big1/raz/GEDMAT/
ReadROI.pl
extract_area.pl
combine_images.pl
gedmat_to_xy.pl
/home/lml/krypton/Corvera/Raz/analyze_gedmat.pl
chres_rpts.pl
filter_rptsinz.pl
examples:
cd /storage/big1/raz/GEDMAT/006_JTH/OM
# generate 006_JTH_OM.areas with all the data (area_mex_border is the number which should replace farea)
# also produce 006_JTH_OM_7masks.i2i and 006_JTH_OM_11masks.i2i so I can check on the validity of the algorithm.
# place diagnostic mask images in /home/lml/krypton/Corvera/Raz/output and the output data file in
# /home/lml/krypton/Corvera/Raz (note: all directories must already exist).
mask_gedmat_images.pl -tmpdir=/home/lml/krypton/Corvera/Raz/output -zip_ending=_Area.zip -results_ending=_Results.txt
-combined_ending=combined.i2i -days=7:11
-save_day=/home/lml/krypton/Corvera/Raz/output
>& /home/lml/krypton/Corvera/Raz/006_JTH_OM.areas
# look at the masks produced:
cd /home/lml/krypton/Corvera/Raz/output
play_results3.pl 006_JTH_OM_day7
or:
fgrep -v # 006_JTH_OM_11masks.play_labels > tmp.play_labels # eliminate the comment line, play doesn't like those.
play -labels tmp.play_labels 006_JTH_OM_7masks.i2i
# compare the full areas produced which what fiji did (as a check). only pull out the good data. they should be very similar:
cd /home/lml/krypton/Corvera/Raz
gedmat_to_xy.pl -cond='if (($date eq "date_sure")&&($perr==0)&&($aerr==0)) {1;} else {0;}'
-xcoord='$area' -ycoord='$farea' 006_JTH_OM.areas | xmgrace -free -pipe
# now grab the data we really want. compare the old way, with the area when the growth half near the well border is masked off:
gedmat_to_xy.pl -cond='if (($date eq "date_sure")&&($perr==0)&&($aerr==0)) {1;} else {0;}'
-xcoord='$area_mex' -ycoord='$area_mex_border' 006_JTH_OM.areas | xmgrace -free -pipe
# it may be hard to see some of these images as they are very large, do it this way (note the -L to keep the -1 boundary values):
addlines _ones.i2i _excluded_region.rpts - | reduceima -bin 3 3 -L - -|play -P -1 R -P -2 G -P -3 B -P -4 C -P -5 Y -
# run with some diagnostics:
cd /storage/big1/raz/GEDMAT/006_JTH/OM
/home/lml/krypton/bin/mask_gedmat_images.pl -tmpdir=/home/lml/krypton/Corvera/Raz -zip_ending=_Area.zip
-results_ending=_Results.txt -combined_ending=combined.i2i -days=7:11
-save_day=/home/lml/krypton/Corvera/Raz/006_JTH_OM_7masks.i2i > /home/lml/krypton/Corvera/Raz/006_JTH_OM.areas
# play.FC14 allows labels to have comment lines (first character of line is a #)
cd home/lml/krypton/Corvera/Raz
play.FC14 -labels 006_JTH_OM_11masks.play_labels 006_JTH_OM_11masks.i2i
chres_rpts.pl -sx=.33 -sy=.33 -truncate -min=1:1 output/_day11.rpts > _day11_bin3.rpts
chres_rpts.pl -sx=.33 -sy=.33 -truncate -min=1:1 output/_day7.rpts > _day7_bin3.rpts
chres_rpts.pl -sx=.33 -sy=.33 -truncate -min=1:1 output/_explant.rpts > _explant_bin3.rpts
reduceima -bin 3 3 output/_day11_noex_halfex.i2i - | addlines - _day11_bin3.rpts - | addlines - _day7_bin3.rpts - |
addlines - _explant_bin3.rpts - | play -P -1 R -
addlines /storage/big1/raz/GEDMAT/006_JTH/OM/D07_20130524_GEDMAT_006_JTH_OM_DAY7_combined.i2i output/_day11.rpts - |
addlines - output/_day7.rpts - | addlines - output/_explant.rpts - | reduceima -bin 3 3 -L - - | play -P -1 R -
# do a bunch of stuff
analyze_gedmat3.pl # apply this perl script to lots of data
cd output
play_results3.pl 006_JTH_OM_day7 # look at some of it
raz_rates.pl # generate stats
source code: in /home/lml/krypton/bin/mask_gedmat_images.pl
mask_gedmat_images4_new.pl
mask_gedmat_images4_new.pl
The easiest way to call this is via analyze_gedmat9.pl, see its help.
This version has the following changes compared with mask_gedmat_images3.pl:
1. @stats = &outline_stats(@outline) was used to calculate explant center (for masking) but
the meanx and meany are just the mean of the outline, it is NOT a center of mass. usually ok for a compact explant.
sub calc_com() is now used instead to calculate the true center of mass.
-use_explant_outline will cause the OLD method to be used instead.
2. bounding box was used to calculate area for sorting the outlines temporally (from smaller to larger area).
but when outlines intersect bounding box can at times NOT be a good surrogate for area.
$roi_area[$r] = ($maxx-$minx)*($maxy-$miny);
handled these two issues by writing sub calc_com().
-use_bbox_area will cause the OLD method to be used instead.
note: fiji areas are already sorted smallest to largest when I first bring them in (extract_area2.pl does that):
$prev_results = `extract_area2.pl -justfiles=$resultsfile`
so I should do not need to use the sorted roi indices on them (so I've been doing this correctly). but the rpts are
being sorted based on bounding box, and fiji areas based on the true area in the Results.txt file. So the ordering
may be different at times. No change was made to this.
mask_gedmat_images4.pl -check_intersect added.
determines when outlines intersect and sets the aerr (area error flag) in the
output file. eventually it will calculate areas a different way when that happens
rather than combining masked images (see ~/krypton/Corvera/Raz/README for 10/18/13). right now only sets the error flag.
Examines all the zip files (produced within fiji when outlining regions) in the current directory. These have
the boundary of the explant, day7 growth, and day11 growth.
note: this program assumes that all the data in the directory is from the same patient and the same tissue type.
It recalculates the area measures by only including that part of the day7 and day11 growth which is between the explant
center and going towards the center of the well; area from the explant center going away from the center of the
well (ie, growing towards the well wall) is excluded. By doing this, we hope to minimize the
effect of growth (as measured by area) being stopped by the well wall when the explant is near the well wall.
Output is written to stdout. The comments there explain the fields. area_mex is the old value, area_mex_border is the new.
This is done by finding all the *_Area.zip files in the current directory, pulling out the outlines in those files
and using that information to recalculate areas. It also needs to find the associated "combined" image file
(*_combined.i2i) to get image size and hence the position of the center of the well) and the fiji results file
(*_results.txt, so the numbers produced here can be easily compared with them).
note: The algorithm assumes that the area NEVER DECREASES with time, and that the explant region is always surrounded
by the day7 region which is surrounded by the day11 region. The latter assumption is sometimes invalid if
the day 11 images were shifted relative to day 7 images; but that is usually ok.
(if necessary, you can manually change the "bad_data" field in the output file to 1 and then use
-cond in gedmat_to_xy.pl to eliminate those stats from further analysis.)
It also assumes the smallest area in each zip file is the original explant area, the next largest area is
called day7 area and the largest area (if 3 are present) is assumed to be day 11 area.
Sometimes when growth is about 0, the outlines on, eg, day7, may have slightly less area than the original
explant, so the outlines might be mis-ordered then. [actually it may read the correct day off the name of
the i2i image now].
note: If only 2 areas (ie, two .roi files) are present in the zip file, they are assumed to be for
the explant and day7 (not day11).
note: This program assumes the center of the combined image (usually 4 well images are combined into one) is the
center of the well. see combine_images.pl. It also assumes the combined image name ends in combined.i2i
(see -combined).
note: some of the scripts I use to then analyze these results will multiply the area by 2, to get a "true" estimate
of growth; since the value produced by mask_gedmat_images3.pl will be (roughly) half of the real growth rates.
usage:
mask_gedmat_images3.pl [options]
options:
-tmpdir=fullpathname directory to use for temporary files which are created, eg: -tmpdir=/home/lml/krypton
-zip_file=file.zip
-zip_file=file.zip:file2.zip:...:filen.zip
only examine the one specified zip file, not all those in the directory
(If the second form is used, you can specify as many as you want.)
note: you may still need to specify -zip_ending so that the ending of this file can be stripped
off correctly (so the -combined_ending and -results_ending can then be added correctly).
-zip_ending=stuff e.g., -zipending=_area.zip . All files which end in this will be analyzed; default = _Area.zip
-skip=file.zip[:file2.zip...] list of zip files (without the full path) to skip.
-results_ending=stuff e.g., -results_ending=_results.txt . This is what the -zip_ending should be replaced by
to get the name of the associated fiji results file. note the _ should be included here
if it was included in -zip_ending.
-combined_ending=stuff e.g., -combined_ending=_combined_reduced_.i2i . default = combined.i2i.
This is any string which helps identify the last part of the combined image filename.
-combined_scaling=#:# e.g., -combined_scaling=2:2 if reduceima -bin 2 2 was applied to get _combined_reduced_.i2i.
-scale=#:# distance and area (eg, -scale=2:4 if reduceima -bin 2 2 was used) scaling. This will multiply
the area stored in the fiji file and the area calculated via my mask by the second number.
The first number will multiply the distance of the explant from the center of the image.
This option is just here to handle 001_BAP and 002_PRO datasets. They were analyzed in fiji
with the images reduced by a factor of 2 in x and y. So the fiji areas produced will be low
by 4x, as will my areas. This option compensates for that. See ~/Corvera/Raz/README for 10/11/13.
-check_intersect check if outlines intersect with the explant's outline. If so, set an error flag on stdout printout
and print info to stderr. eventually will just use area numbers rather than masking area in this situation.
that is not done yet ...
-save_day[=directory]
saves the final binary mask for each days growth (eg, day7 growth minus the explant and minus the part growing towards the wall)
into a 3d mask image (patientid_tissue_day#_mask.i2i). This image will be reduced down in xdim and ydim from the
original. patientid_tissue_day#_.play_labels will also be created (labels which well each z slice comes from).
This is mostly for quality control purposes.
In addition, patientid_tissue_day#.rpts will be created (similarly reduced down in size) (coords may be off by 1?).
View the file via: addlines maskim.i2i maskim.rpts - |play.FC14 -P -1 R -labels maskim.play_labels -
or via: play_results3.pl
patientid_tissue_day#_orig.i2i will be the original images (also reduced in xdim and ydim, and only one 2d slice
from each original 3d image).
note: older versions of play may need the comment line at the top of maskim.play_labels removed.
note: if you want images to be in another directory, specify a full pathname, eg -save_day=/home/lml/krypton (without last /).
note: any existing file with these names will be OVER-WRITTEN.
note: this option creates lots of temporary files (in tmpdir) of the form __*.i2i, any existing
files that match this will be DELETED.
see also: montage
-patient5 patient5 files have some odd naming. use this option if analyzing them.
-use_explant_outline legacy. specify this if you want the center of the explant to be based on the average of the pixel positions
of just the pixels along the boundary (rather than the center of mass).
-use_bbox_area legacy. specify this if you want to order the outlines based on their bounding box area rather than their true area.
You should probably specify both options (if you want either one), I'm not sure about consistency if only one is specified.
-just_check just check whether each zip roi outline has the correct number of associated image files.
eg, if the zip file has 3 outlines (explant, firstday, secondday) there should be two i2i files with DAY# in their name.
prints to stderr info about those which don't have the correct number. does nothing else.
-just_stats goes a bit further than -just_check (does that too), sets -verbose and prints out stats about each outline. then exits.
-verbose some extra info to stdout
-debug prints some tracing info to stderr
-print_fname print name of each zip file as it is being analyzed, to stderr.
-dlevel=# extra debugging. the bigger the number the more the printout. # is an integer starting at 1.
-dlevel=1 will print a trace of commands to execute to mimic (parts of) this perl script.
-noclean don't remove the temporary files which were created
the files are: _ones.i2i _explant.rpts _day11.rpts _day7.rpts _excluded_region.rpts
_explant.i2i _day11.i2i _day7.i2i _day7_noex.i2i _day11_noex.i2i _day7_noex_halfex.i2i _day11_noex_halfex.i2i
(if multiple zip files were analyzed, only the files from the last one will be left, since they get reused)
-dont_recalc only for diagnostics. does NOT restrict the analysis to just the growth away from the wall.
with this specified this entire program should just reproduce the numbers we already
have in *Results.txt since that uses the entire area. ie, this make the program a no-op.
see also:
/home/lml/krypton/Corvera/Raz/README
analyze_gedmat3.pl a script which calls mask_gedmat_images3.pl for all of Raz's data. Also versions 4,5,6, and 7.
play_results3.pl
raz_rates.pl
mask_gedmat_images.pl
mask_gedmat_images2.pl
/storage/big1/raz/GEDMAT/
ReadROI.pl
extract_area.pl
combine_images.pl
gedmat_to_xy.pl
/home/lml/krypton/Corvera/Raz/analyze_gedmat.pl
chres_rpts.pl
filter_rptsinz.pl
examples:
cd /storage/big1/raz/GEDMAT/006_JTH/OM
# generate 006_JTH_OM.areas with all the data (area_mex_border is the number which should replace farea)
# also produce 006_JTH_OM_7masks.i2i and 006_JTH_OM_11masks.i2i so I can check on the validity of the algorithm.
# place diagnostic mask images in /home/lml/krypton/Corvera/Raz/output and the output data file in
# /home/lml/krypton/Corvera/Raz (note: all directories must already exist).
mask_gedmat_images.pl -tmpdir=/home/lml/krypton/Corvera/Raz/output -zip_ending=_Area.zip -results_ending=_Results.txt
-combined_ending=combined.i2i -days=7:11
-save_day=/home/lml/krypton/Corvera/Raz/output
>& /home/lml/krypton/Corvera/Raz/006_JTH_OM.areas
# look at the masks produced:
cd /home/lml/krypton/Corvera/Raz/output
play_results3.pl 006_JTH_OM_day7
or:
fgrep -v # 006_JTH_OM_11masks.play_labels > tmp.play_labels # eliminate the comment line, play doesn't like those.
play -labels tmp.play_labels 006_JTH_OM_7masks.i2i
# compare the full areas produced which what fiji did (as a check). only pull out the good data. they should be very similar:
cd /home/lml/krypton/Corvera/Raz
gedmat_to_xy.pl -cond='if (($date eq "date_sure")&&($perr==0)&&($aerr==0)) {1;} else {0;}'
-xcoord='$area' -ycoord='$farea' 006_JTH_OM.areas | xmgrace -free -pipe
# now grab the data we really want. compare the old way, with the area when the growth half near the well border is masked off:
gedmat_to_xy.pl -cond='if (($date eq "date_sure")&&($perr==0)&&($aerr==0)) {1;} else {0;}'
-xcoord='$area_mex' -ycoord='$area_mex_border' 006_JTH_OM.areas | xmgrace -free -pipe
# it may be hard to see some of these images as they are very large, do it this way (note the -L to keep the -1 boundary values):
addlines _ones.i2i _excluded_region.rpts - | reduceima -bin 3 3 -L - -|play -P -1 R -P -2 G -P -3 B -P -4 C -P -5 Y -
# run with some diagnostics:
cd /storage/big1/raz/GEDMAT/006_JTH/OM
/home/lml/krypton/bin/mask_gedmat_images.pl -tmpdir=/home/lml/krypton/Corvera/Raz -zip_ending=_Area.zip
-results_ending=_Results.txt -combined_ending=combined.i2i -days=7:11
-save_day=/home/lml/krypton/Corvera/Raz/006_JTH_OM_7masks.i2i > /home/lml/krypton/Corvera/Raz/006_JTH_OM.areas
# play.FC14 allows labels to have comment lines (first character of line is a #)
cd home/lml/krypton/Corvera/Raz
play.FC14 -labels 006_JTH_OM_11masks.play_labels 006_JTH_OM_11masks.i2i
chres_rpts.pl -sx=.33 -sy=.33 -truncate -min=1:1 output/_day11.rpts > _day11_bin3.rpts
chres_rpts.pl -sx=.33 -sy=.33 -truncate -min=1:1 output/_day7.rpts > _day7_bin3.rpts
chres_rpts.pl -sx=.33 -sy=.33 -truncate -min=1:1 output/_explant.rpts > _explant_bin3.rpts
reduceima -bin 3 3 output/_day11_noex_halfex.i2i - | addlines - _day11_bin3.rpts - | addlines - _day7_bin3.rpts - |
addlines - _explant_bin3.rpts - | play -P -1 R -
addlines /storage/big1/raz/GEDMAT/006_JTH/OM/D07_20130524_GEDMAT_006_JTH_OM_DAY7_combined.i2i output/_day11.rpts - |
addlines - output/_day7.rpts - | addlines - output/_explant.rpts - | reduceima -bin 3 3 -L - - | play -P -1 R -
# do a bunch of stuff
analyze_gedmat3.pl # apply this perl script to lots of data
cd output
play_results3.pl 006_JTH_OM_day7 # look at some of it
raz_rates.pl # generate stats
source code: in /home/lml/krypton/bin/mask_gedmat_images.pl
membrane3D.pl
A script to run the routines necessary to perform 3D membrane segmentation.
note: it needs write permission in the directory where inputimage.i2i resides since all output images go there too.
It will run the following programs, in order:
1. /home/lml/krypton/facil/i2i2mhd (convert to mhd format)
2. /home/lml/krypton/segmentation/MR-Source/ACME/ACME-Binary/bin/cellPreprocess (median filter, fill holes)
3. /home/lml/krypton/segmentation/MR-Source/ACME/ACME-Binary/bin/resample (resample pixel sizes)
4. /home/lml/krypton/segmentation/MR-Source/ACME/ACME-Binary/bin/multiscalePlateMeasureImageFilter (determine the planarity of each region)
5. /home/lml/krypton/segmentation/MR-Source/ACME/ACME-Binary/bin/membraneVotingField3D (update planarity based on neighbors voting)
6. /home/lml/krypton/segmentation/MR-Source/ACME/ACME-Binary/bin/membraneSegmentation (produce segmented regions via a watershed algorithm)
Each program expects input files of a certain type (e.g. unsigned char).
usage:
membrane3D.pl [options] inputimage.[i2i|mhd|mha] outimage.mhd
output files:
inputimage.mhd produced by i2i2mhd - short ints
inputimage_med.mhd produced by cellPreprocess
inputimage_med_resamp.mhd produced by resample, unsigned char
inputimage_med_resamp_planar.mhd produced by multiscalePlateMeasureImageFilter, floating pt (doubles) between 0 and 1, how "planar" a pt is.
inputimage_med_resamp_eigen.mhd produced by multiscalePlateMeasureImageFilter, an eigenmatrix at each pt
inputimage_med_resamp_eigen_voting.mhd produced by membraneVotingField3D, result of voting, unsigned char
outimage.mhd produced by membraneSegmentation, result of watershed, each region labeled with an id (short integer).
note: all units below are in the same units as the pixel units (I think). So if pixel spacing is 2 (um) in x, and a radius
of 2 is specified, that is a one pixel radius.
step 1. options for converting i2i to mhd:
-convert=dx:dy:dz The first step is converting our i2i format to mhd format. This specifies the pixel spacing
in x y and z directions for that. default assumes 1:1:1. The "conversion" really just creates inputimage.mhd which
is a text header which points to the original i2i image and gives some info about it.
If the input image has a .mhd or .mha extension already (instead of .i2i) then no conversion is done.
Olga's images acquired with a 10x objective have a pixel size of .645 um/pixel (and z spacing of 10 um).
Example: G03_x_1_1_binned.i2i has a pixel size of 2.5x2.5x10. So -convert=2.5:2.5:10 specifies this in the mhd header.
step 2. options for cellPreproces:
this may produce an unsigned char image (?)
-median=# radius of median filtering preprocessing (and hole filling). If this is not specified it isn't done.
step 3. options for resampling:
this may produce an unsigned char image (?)
-resample=x:y:z resample in the x y and z directions, eg. -resample=2:2:1 will halve the x and y dimensions and leave z unchanged.
note: this will change the spacing between the pixels; but the routines which follow will understand that.
For example: if after resampling, the pixels are spaced 10 um apart, then -planar=10 will use a sigma of 1 pixel.
If this is not specified it isn't done.
note: it may be necessary to do the above operations, which also convert the image into unsigned char, which is required for
step 4, multiscalePlateMeasureImageFilter?
note: step 6 (watershed) may also work best if the pixel sizes are the same in all directions.
step 4. options relating to planarity:
the planarity image produced will be doubles from 0-1.
-planar=# the sigma of the Gaussian used when classifying pixels as planar or not. default = 1
-alpha=# see the help for /home/lml/krypton/segmentation/MR-Source/ACME/ACME-Binary/bin/multiscalePlateMeasureImageFilter
-beta=# ditto
-gamma=# ditto
-c=# ditto
-planaropts="options for multiscalePlateMeasureImageFilter here"
available options (so far, but anything specified here will be passed), : -a # -b # -g # -c # -d -s -B -T
options which go here should be specified in the format that multiscalePlateMeasureImageFilter wants them,
for example: -planaropts="-c 5 -d -B"
not in the alternative format that Membrane3D.pl desires (e.g. -planaropts="-a 5" is correct, -planaropts="-alpha=5" is wrong).
-stop_after_planar don't do anything after this step (e.g. voting for planar patches may not make sense if -B or -T specified)
step 5. options relating to voting:
the resulting tensor vote image will be unsigned chars from 0-255.
-region=# the sigma which defines the region within which a pixel influences other pixels. default = 10
step 6. options relating to watershed segmentation:
The resulting image will be long ints (id numbers) of regions.
See the help for /home/lml/krypton/segmentation/MR-Source/ACME/ACME-Binary/bin/membraneSegmentation
-thresh=# threshold value. this is applied to the tensor voting image to turn it into a binary image. then a distance
transform and watershed are applied. default = 128
-segopts="options for membraneSegmentation here"
for example: -segopts="-S -l 20 -a 5 -c -D" will use this exact string as options.
options so far: -S -l # -a # -s # -n -i # -w # -m # -c -v -d -D
other options:
-verbose echo what is being done to stderr
-dummy don't really do anything; echo what would have been done to stderr.
example:
reduceima -bin 4 4 G03_x_1_1.i2i G03_x_1_1_binned.i2i - go from .645um/pixel, 10um in z -> 2.5um/pixel, 10um in z
# -convert: just put correct pixel size info into an mhd header
# -median: smooth with a filter 10 um (4 pixels, since they are now 2.5um/pixel) in radius (diameter?)
# -resample: change to a square pixel (10 um in x y and z). Change image format to unsigned char (needed by MultiscalePlateMeasureImageFilter)
# -planaropts: options for planarity filter
# -region: width (in um) of neighbors for voting on planarity and updating it.
# -segopts:
# -thresh: threshold for watershed segmentation
membrane3D.pl -convert=2.5:2.5:10 -median=10 -resample=2:2:1 -planaropts=" " -region=50 -segopts=" " -thresh
G03_x_1_1_binned.i2i G03_x_1_1_results.mhd
see also: /home/lml/krypton/segmentation/MR-Source/ACME/lmltest/README
/home/lml/krypton/segmentation/FindingCellMembranesIn3D.pdf
i2i2mhd - convert i2i into mhd format
mhd2i2i - convert an mhd image (but not an eigenimage)
RegionId2Mesh - will convert the ID image produced into a mesh image or rpts file
mayavi2 - to visualize *.mha and *.mhd files.
Fiji - my version has a File-> Import metaimage plugin
Norio_stats.pl
This programs takes each pts file in the current directory, thresholds the associated vinculin image, applies that mask to the trip6 image, and produces a .coloc file from the masked trip6 image (using mask_and_coloc.pl); overlap statistics are then calculated (and put into a file name *_masked.coloc where * is the root of the trip6 image name) and finally the highest ratio data from each image pair is printed to stdout (using sort_overlap3.pl). stats.pl [options] > stats.out -nothing : do nothing, just echo what would be done -h : print this help -thresh=# : threshold for the vinculin image. default = 20 -regions=#:# : only allow mask regions within the specified min:max size (in pixels, inclusive). : Also creates *_count.i2i and *.countobjs (* = root of vinculin image name) intermediate files. : note: these counts are for the vinculin image after the mask (pts file) is applied. -clean: : clean up (ie, remove) *_count.i2i and *.countobjs images (if -regions specified)
run2_Ronghua.pl
this program goes through Ronghua's directory (see -dir option) analyzing files. output is put into the current working directory (ie, the directiory run.pl is run from). A tag in the name of the form Ryr_thresh_minsize_maxsize_Bk_thresh_minsize_maxsize is added to the output name of name.patchdist (nearest voxel distance between whole patches) and name.comdist (distances between nearest center of mass of the patches). usage: run.pl [options] options: -dir=directorypath : without the final slash. directory to analyze. default = /mnt/big3/rzg/mtc/immunocytochemistry/july05/imcc/70105/epr -ryrthresh=# : threshold for ryr (488) images, default = -min_ryrsize=# : minimum allowed object size in pixels, default = 5 -max_ryrsize=# : max allowed object size in pixels, default = 150 -min_ryriod=# : minimum allowed object iod, default = 354 -max_ryriod=# : maximum allowed object iod, default = 10000000 -bkthresh=# : threshold for bk (594) images, default = -min_bksize=# : minimum allowed object size in pixels, default = 5 -max_bksize=# : max allowed object size in pixels, default = 50 -min_bkiod=# : minimum allowed object iod, default = 4686 -max_bkiod=# : maximum allowed object iod, default = 10000000 -dx=# : relative distance in x, default = 1 -dy=# : relative distance in y, default = 3 -dz=# : relative distance in z, default = 1 -noclean -debug -verbose
countobjs
Usage:
countobjs [options] outputfile image1.i2i [image2.i2i]
Description:
Count objects in co-localized images. If only one image given
the objects will be counted in that one image. Copious amounts
of information will be written to the output file.
A - for outputfile will write to stdout.
outpufile can be analyzed with objs2bb.pl and objs2dist.pl
Options:
-C # # : set opacity and brightness level to these values (0-255 only)
-k : use original images for data analysis (so scale and black level not important and
-t option can be in same units as original, unscaled, image values)
-S # # # : image # scale and black level (default autoscale image)
-r # # # # # # : region of image to count
(-r x0 y0 z0 x1 y1 z1)
( ^lower left ^upper right)
(zero indexed coordinates)
(default whole image)
-s # # : size of smallest and largest objects to count
(default 1-100000)
-m # # : min and max iod of allowable objects (applied to image1 only).
-t thresh1 thresh2 : thresholds (applied after -S or default scaling, unless -k specified).
Like left brightness slider in DAVE.
if only one image, second number still required, but ignored.
-o <image.i2i> : output image with all objects represented by the voxel
with the maximum intensity in the object
the intensity of the voxel will be the IOD of the object.
-O <image.i2i> : output image with all pixels in each object having
the intensity of the original objects IOD.
NOTE: this image will be in floating point format.
use float2int to convert it back to "normal" format.
-Q <image.i2i> : output image with all pixels in each object having
a unique id # (1-# of objects). Must be < 32766 objects.
NOTE: this image will be in standard signed short int format.
-M # : When countobjs is given 2 images, this flag will
turn of some subset of the voxels for colcalization.
calculations.
0: turn off colcalized voxels (default on)
1: turn off image 1 voxels (default on)
2: turn off image 2 voxels (default on)
-q : quiet. don't print out all objects, but just summary statistics
-Z : simple analysis, no sliders, opacity,etc. can use with -t option.
-h : show this message.
-H : show even more help.
Notes:
A - in place of an image name means stdin or stdout.
Source code in /storage/big1/lml/jac/was_invitro/Projects/countobjs
countobjsmax3d
Usage:
countobjsmax3d [options] -Q imageid.i2i outputfile image1.i2i
Description:
This version of countobjs finds 3d maxima in image1.i2i, then finds all pixels connected
to each maxima and downhill from it. These are labelled with a unique id number and stored
in imageid.i2i (specified with the -Q option, which is REQUIRED for this version)
You may want to display imageid.i2i after using colorize_image on it.
Not tested yet.
Count objects in co-localized images. If only one image given
the objects will be counted in that one image. Copious amounts
of information will be written to the output file.
A - for outputfile will write to stdout.
outpufile can be analyzed with objs2bb.pl and objs2dist.pl
Options:
-r # # # # # # : region of image to count
(-r x0 y0 z0 x1 y1 z1)
( ^lower left ^upper right)
(zero indexed coordinates)
(default whole image)
-s # # : size of smallest and largest objects to count
(default 1-100000)
-m # # : min and max iod of allowable objects (applied to image1 only).
-t thresh1 thresh2 : thresholds (applied after -S). Like left brightness slider in DAVE.
if only one image, second number still required, but ignored.
-o <image.i2i> : output image with all objects represented by the voxel
with the maximum intensity in the object
the intensity of the voxel will be the IOD of the object.
-O <image.i2i> : output image with all pixels in each object having
the intensity of the original objects IOD.
NOTE: this image will be in floating point format.
use float2int to convert it back to "normal" format.
-Q <image.i2i> : output image with all pixels in each object having
a unique id # (1-# of objects). Must be < 32766 objects.
NOTE: this image will be in standard signed short int format.
-M # : When countobjs is given 2 images, this flag will
turn of some subset of the voxels for colcalization.
calculations.
0: turn off colcalized voxels (default on)
1: turn off image 1 voxels (default on)
2: turn off image 2 voxels (default on)
-q : quiet. don't print out all objects, but just summary statistics
-Z : simple analysis, no sliders, opacity,etc. can use with -t option.
-h : show this message.
-H : show even more help.
Notes:
A - in place of an image name means stdin or stdout.
Source code in /storage/big1/lml/jac/was_invitro/Projects/countobjs
threshold.pl
threshold.pl -numvox=# [-produce=newimage.i2i] image.i2i calculate the largest threshold for image.i2i which has at least # voxels equal or above it. Prints the threshold and the number above threshold to stdout. options: -produce=newimage.i2i : take the calculated threshold, apply it to image.i2i to produce newimage.i2i
thresholdz.pl
thresholdz.pl -numvox=# [-produce=newimage.i2i] image.i2i calculate the largest threshold for image.i2i which has at least # voxels equal or above it. Prints the threshold and the number above threshold to stdout. options: -produce=newimage.i2i : take the calculated threshold, apply it to image.i2i to produce newimage.i2i
2Ddistances
Calculates a histogram of the amount of light as a function of distance from boundary (0 valued) voxels as specified in the mask. Applies a 2D mask to a 2D time series. Usage: 2Dhistogram [options] inimage.i2i inmask.i2i Options: -v: verbose Source code in ~krypton/lml/facil -d dimage.i2i : save the distance image as dimage.i2i (converted to ints by casting) -n: don't normalize the histogram by the number of voxels in the mask at that distance -a #: average together this many time pts for output histogram. default = 20 see also: histogramL, histogram2 Copyright 2010 University of Massachusetts Medical School and the Biomedical Imaging Group All rights reserved. Explicit permission to use this program must be received prior to use.
analyze_histo
# /home/lml/krypton/bin/analyze_histo Analyzes the output histogram image produced by histogram2 via the -f option. Prints out 2 columns: first column is bin number, second column is value at the cutoff. This is similar to the program "threshold" but instead of one threshold value for the entire image, it gives one at each distance (from a membrane). Usage: analyze_histo [options] aimage_H.i2i options: -v: calculate cutoff based on percentage of voxels instead of percentage of light -f #: fractional cutoff (e.g., .80 = 80%), default = 0.950000 -w #: number of bins for running average (default = 5), should be odd. -t #: only look at voxels >= intensity (must be >= 0), default = 1 Examples: histogram2 -f a_H.i2i -O a_H.histo -o -p 0 0 0 -d 40 -z 3 mask.i2i a.i2i b.i2i analyze_histo -f .95 a_H.i2i > a_H.cutoffs See also: histogram, histogram2, threshold source code in /storage/big1/lml/jac/was_invitro/Projects/boundary/boundary
analyze_yaodl
This program reads in a yaodl file as produced by DAVE (volume widget menu: volume to surface ...)
(hmc produces an ascii format which analyze_yaodl can't handle).
It identifies connected surfaces (since DAVE considers all surfaces from one pass as one object).
It then calculates and prints the volume and surface area of each connected surface. Optionally
it will write out a yaodl files (-y) for the largest surface found for each object, with better normals.
This surface can be read back into DAVE with -y option.
Usage: analyze_yaodl [options] infile.ydl
options:
-y largest.ydl: write out largest surface of eac object to largest.ydl
-i image.i2i: read in the image file associated with infile.ydl (not needed for anything yet).
-n: recalculate "better" normals (use this with the -y option)
(dave used to produce terrible normals, now they are ok, so this isn't needed anymore)
-v: verbose
note: source code in /storage/big1/lml/Fischer/src directory.
BestPath
Finds the brightest path from a start pixel to a stop pixel (1-indexed). Usage: BestPath [options] image.i2i Options: -t #: threshold. Only pixels with values > # are allowed in the path, default = 0.000000 -s x y z: start coordinate (1-indexed). default = (1,1,1) -S x y z: stop coordinate (1-indexed). default = (1,1,1) -v: verbose Source code in ~krypton/lml/facil
BestPath3D
Finds the brightest path from a start pixel to a stop pixel (1-indexed).
Converts voxels into nodes in a graph and finds the lowest cost path from the start node to the stop node.
The cost of each node is (maxval-imval), so bright nodes have less cost. Output in rpts format to stdout.
BestPath3D always turns all (above threshold) voxels in the image into graph nodes. BestPath3D_ellipse only
looks at voxels within an ellipse with a major axis connecting the start and endpts.
Usage:
BestPath3D [options] image.i2i > image.rpts
BestPath3D_ellipse [options] image.i2i > image.rpts
Options:
-t #: threshold. Only pixels with values > # are allowed in the path, default = 0.000000
-s x y z: start coordinate (1-indexed).
-S x y z: stop coordinate (1-indexed).
Up to 1000 pairs of start/stop pixels can be specified. At least one pair must be specified.
-r scale black rescale image values so new = (old-black)/scale prior to being used
-n name name of object that is placed into the rpts file.
-f exfac perpfac: Only used if you are running BestPath3D_ellipse
sets principal axis a to have the specified endpts (from -s and -S), then expands it by factor exfac,
and sets the length of the perpendicular b and c axes to be perpfac times the original axis a length
defaults: exfac = 1.000000, perpfac = 1.000000
-v: verbose
-d: debug
-D debug.i2i: debug. also write out image with 50 added to all voxels within the ellipsoid.
Examples:
make_spiralim -d 200 200 30 -r 70 -p spiral_small.i2i # create a test image
-s 171 101 1 -S 170 99 30 # written out by make_spiralim
BestPath3D_ellipse -v -t -1 -s 171 101 1 -S 170 99 30 -f 3 13 spiral_small.i2i > spiral.rpts
note: spiral_small.i2i is a tough case, since the spiral goes very far from the start/stop points
This requires a -f 3 to expand out the endpts and then a 13 to expand out perpendicularly.
If you just expand out perpendicularly (e.g. -f 1 13) the ellipsoid doesn't grow faster enough
near the endpts so some of the spiral data is not included in the ellipsoid. See -D option.
See also: make_spiralim, BestPath, BestPath2, make_ellipsoid, DAVE -> Stuff -> Best Path ...
Source code in ~krypton/lml/facil
BestPath3D_ellipse
Finds the brightest path from a start pixel to a stop pixel (1-indexed).
Converts voxels into nodes in a graph and finds the lowest cost path from the start node to the stop node.
The cost of each node is (maxval-imval), so bright nodes have less cost. Output in rpts format to stdout.
BestPath3D always turns all (above threshold) voxels in the image into graph nodes. BestPath3D_ellipse only
looks at voxels within an ellipse with a major axis connecting the start and endpts.
Usage:
BestPath3D [options] image.i2i > image.rpts
BestPath3D_ellipse [options] image.i2i > image.rpts
Options:
-t #: threshold. Only pixels with values > # are allowed in the path, default = 0.000000
-s x y z: start coordinate (1-indexed).
-S x y z: stop coordinate (1-indexed).
Up to 1000 pairs of start/stop pixels can be specified. At least one pair must be specified.
-r scale black rescale image values so new = (old-black)/scale prior to being used
-n name name of object that is placed into the rpts file.
-f exfac perpfac: Only used if you are running BestPath3D_ellipse
sets principal axis a to have the specified endpts (from -s and -S), then expands it by factor exfac,
and sets the length of the perpendicular b and c axes to be perpfac times the original axis a length
defaults: exfac = 1.000000, perpfac = 1.000000
-v: verbose
-d: debug
-D debug.i2i: debug. also write out image with 50 added to all voxels within the ellipsoid.
Examples:
make_spiralim -d 200 200 30 -r 70 -p spiral_small.i2i # create a test image
-s 171 101 1 -S 170 99 30 # written out by make_spiralim
BestPath3D_ellipse -v -t -1 -s 171 101 1 -S 170 99 30 -f 3 13 spiral_small.i2i > spiral.rpts
note: spiral_small.i2i is a tough case, since the spiral goes very far from the start/stop points
This requires a -f 3 to expand out the endpts and then a 13 to expand out perpendicularly.
If you just expand out perpendicularly (e.g. -f 1 13) the ellipsoid doesn't grow faster enough
near the endpts so some of the spiral data is not included in the ellipsoid. See -D option.
See also: make_spiralim, BestPath, BestPath2, make_ellipsoid, DAVE -> Stuff -> Best Path ...
Source code in ~krypton/lml/facil
boundaryL
Usage:
boundary [options] original.i2i
Description:
Boundary takes an image, applys a threshold to the image,
rolls a ball outside then inside the object. This defines
what pixels are considered inside, outside, and boundary.
Expects: Input image and optionally some given options
Modifies: Nothing
Returns: Output image with the name of image with _B added.
(if input.i2i was original image input_B.i2i
is output image)
Options:
-I : Do not roll ball inside of the cell.
(rolling the ball inside the cell results
in a smoother surface)
-r # : Set radius to # for sphere exterior to cell.
(default = 3)
-R # : Set radius to # for sphere interior to cell.
(default = 3)
-t # : Set threshold for all z slices to > #.
(default is 10)
-T filename: Use file filename for thresholds for each z slice.
(one threshold for each zslice - one threshold on a line)
-X : Restrict search in X direction.
(can be used to cap a cylinder whose long axis
is along the x axis)
-Y : Restrict search in Y direction.
(can be used to cap a cylinder whose long axis
is along the y axis)
-i string : Add string to history of output image.
-h : Print this message.
-H : Print this message and then some.
Caveats:
Requires lots of memory and time to run (megabytes+minutes).
Notes:
Entire image name (with extension) is required.
Pixel values: boundary = 2, outside = 4, inside = 6.
Output image is padded on all sides by 2*radius+1 of
sphere rolled on the outside of the object.
Keywords:
surface location, boundary, boundaries, border
Additional programs:
histogram, shrink_wrap, createBoundary, analysis
Last Changed: Wed Feb 10, 1993
calc_area
This program takes in a surface image and a data image and calculates: 1). the area of the surface (number of nonzero voxels) 2). the number of data voxels (nonzero voxels in image2) 3). the number of surface voxels (area) within a specified distance of a data voxel. This information is printed to stderr. Typically one might apply a shrink_wrap to, say, a Vinculin image of a dual labelled image pair. Then make_surface would convert this to a surface. After this, map_to_surface would be applied to the 2 dual labelled images. Then overlap would be used to calculate the overlap of the two data sets (given some overlap range, which defaults to 1). Then this program (calc_area) is used to get the area of the surface and the amount of it which is examined using the range specified to the overlap program. This is important since to calculate the probability that the overlap was due to chance you want to know what percentage of the area was included in the overlap calculation. Usage: calc_area [options] surface.i2i data.i2i options: -r range: range in voxels (default = 1). -i image.i2i: also produce an output image this image will have bit 2 set for original data voxels and bit 1 set for surface voxels near (or on) data voxels (so it would be a voxel with a value of 1 or 3). note: entire image name (with extension) is NOT required. note: a - in place of an image name means stdin or stdout. note: a compressed image will be automatically be uncompressed. Source code in /storage/big1/lml/was_invitro/Moore/pkc
closest_object
Calculates the closest distance from an object in image1 to an object in image2. Also calculates closest distance
between objects within each image separately. Prints out info about matches. If no other object is within the
maximum distance (see -m option), the object is printed matching to object -1 at a huge distance.
Histogram of these distances is printed to stdout.
The input images should have each pixel in an object having the object id as its value. These images can be
produced with "countobjs -Q" or "objs_via_ascent"
Usage:
closest_object [options] image1.i2i image2.i2i > im1_im2.hist
Options:
-t # #: image1 and image2 thresholds, pixels > # are above threshold, default = 0.
-w # # #: x,y, zweights for pixel distances (ie, pixel size), default = 1 for each.
-m # : maxdistance (in units used for -w) to look for the closest pixel, default = 10.000000
-r #: resolution for output histogram, ie, binsize, default = 0.100000
-e lowx lowy lowz hix hiy hiz: only look at voxels within this extent (subvolume), 1-indexed, inclusive.
-s minsize maxsize: only look at objects in image1.i2i within the size range (inclusive, in voxels)
-S minsize maxsize: only look at objects in image2.i2i within the size range (inclusive, in voxels)
-i miniod maxiod: only look at objects in image1.i2i within the iod range (inclusive)
-I miniod maxiod: only look at objects in image2.i2i within the iod range (inclusive)
-d image1d.i2i: the original intensity image for image1 (since image1 has pixel values as id numbers)
-D image2d.i2i: the original intensity image for image2 (since image2 has pixel values as id numbers)
if you want the iod or pixel intensity stats to be meaningful, you need to provide
the original data by using the -d and -D options.
-v: verbose
-V: very verbose
Examples:
Find the distance from the closest Ryr voxel in each object to a BK object. Record the iod
of the BK object in the histogram:
countobjs -t 5 10 -Q ryr_id.i2i ryr.countobjs ryr.i2i : this finds Ryr connected objects and labels them with id
countobjs -t 10 10 -O bk_iodR.i2i bk.countobjs bk.i2i : this calculates the iod of BK objects
float2intL -M 30000 bk_iodR.i2i bk_iod.i2i : rescale BK iod to integer format
closest_object -w 1 1 3 -o 5 100 ryr_id.i2i bk_iod.i2i > ryr_bk.info
cull -s -c -1 -4 < ryr_bk.info |xmgrace -source stdin
note: you may want to take a 10 pt running avg inside xmgrace to compensate for
issues related to discretization of distances
See also:
closest_voxel objs_via_ascent /home/lml/krypton/Zhuge/BKandRyr/graph.pl,match.pl
Bugs:
Source code in ~krypton/lml/facil
closest_voxel
Calculates the closest distance from an above threshold voxel in image1 to an above threshold voxel in image2.
Histogram of these distances is printed to stdout.
Usage:
closest_voxel [options] image1.i2i image2.i2i > im1_im2.hist
Options:
-t # #: image1 and image2 thresholds, pixels > # are above threshold, default = 0.
-w # # #: x,y, zweights for pixel distances (ie, pixel size), default = 1 for each.
-m # : maxdistance (in units used for -w) to look for the closest pixel, default = 10.000000
-r #: resolution for output histogram, ie, binsize, default = 0.100000
-s lowx lowy lowz hix hiy hiz: only look at voxels within this subvolume, 1-indexed, inclusive.
-Q: only the voxel in each image1 object closest to an image2 voxel should count.
Use this if image1.i2i was created via the -Q option from countobjs.
-o minsize maxsize: only look at objects in image1.i2i within the size range (inclusive, in voxels)
must use -Q option along with this.
-p: also print the detailed info of which voxel matched to which other voxel
-v: verbose
-V: very verbose
Examples:
Find the distance from the closest Ryr voxel in each object to a BK object. Record the iod
of the BK object in the histogram:
countobjs -t 5 10 -Q ryr_id.i2i ryr.countobjs ryr.i2i : this finds Ryr connected objects and labels them with id
countobjs -t 10 10 -O bk_iodR.i2i bk.countobjs bk.i2i : this calculates the iod of BK objects
float2intL -M 30000 bk_iodR.i2i bk_iod.i2i : rescale BK iod to integer format
closest_voxel -w 1 1 3 -o 5 100 -Q ryr_id.i2i bk_iod.i2i > ryr_bk.info
cull -s -c -1 -4 < ryr_bk.info |xmgrace -source stdin
note: you may want to take a 10 pt running avg inside xmgrace to compensate for
issues related to discretization of distances
See also:
graph.pl
Bugs:
Source code in ~krypton/lml/facil
coloc
This program reads in 2 or 3 images, and writes out an image in which colocalized voxels (voxels > threshold in all images) are kept, others set to 0. By default colocalized voxels are set to image1 values. Usage: coloc [options] image1 image2 [image3] outimage options: -a #: threshold for image 1 (default = 0) -b #: threshold for image 2 (default = 0) -c #: threshold for image 3 (default = 0) -va: make outimage pixel value be value of image 1 -vb: make outimage pixel value be value of image 2 -vc: make outimage pixel value be value of image 3 -vu #: make outimage pixel value be user specified, # note: source code in /storage/big1/lml/Laxman/src/coloc.c
coloc3ims
This programs reads in three images and produces a new version of image3. Each voxel which is colocalized (has image1 > thresh1 and image2 > thresh2) has the value in image3 set to 32000 (-n 32000). Otherwise the value in image3 is not changed. Once these modifications are made, image1.i2i image2.i2i and newimage3.i2i can be read into DAVE (in that order) and voxels which are colocalized in the first two channels will be guaranteed to be colocalized in the third channel. Usage: coloc3ims [options] image1.i2i image2.i2i image3.i2i newimage3.i2i options: -t thresh1 thresh2: thresholds for image1 and image2, defaults = 0 and 0 -n newval: instead of setting newimage3 to 0, set it to newval -m: instead of setting newimage3 to 0, set it to the max value in image3.i2i note: entire image name (with extension) is NOT required. note: a - in place of an image name means stdin or stdout. note: a compressed image will be automatically be uncompressed.
coloc_tseries2
Similar to coloc_tseries, but compares a 2D time series to itself.
Prints xmgr formatted graphs to stdout.
Usage: coloc_tseries2 image1 outfile
options:
-t thresh: only count voxels > thresh. default = 0
-z start stop: only look at these z planes (inclusive, 1-indexed). default = all z planes.
-n: don't include a legend in the stdout.
-v: verbose
source code in: /home/lml/krypton/facil
see also: /home/lml/krypton/Corvera/paper2010/Dynamics_in_Dave/README
com_2d_objs
This program takes an image and finds the intensity weighted center of mass of each 2d object.
Object 1 is all pixels with value = 1, object 2 is pixels with value 2, etc. Max objects = 10
Statistics are written to stdout
Usage: com_2d_objs [options] image.i2i
options:
-z start stop: zplanes to analyze. 0-indexed inclusive. default is all z planes
-r #: put data out rpts format (integer coords, only center of mass info). Only use if only one object.
# is the pixel value (!= 0), usually -1 through -7.
-o: coords should be one-indexed (ie, min x = 1). default = 0. -r sets this automatically.
-v: verbose
Source in /home/lml/krypton/facil
See Also: /home/lml/krypton/Lawson/Jacques/README
Examples:
planimeter3.20081007 -I 110215ligand_0_491.i2i -R 110215ligand_0_491_cell1.rpts
interp -S -p -1 -o cell1 110215ligand_0_491_cell1.rpts 110215ligand_0_491_cell1_interpS.rpts
create_dcimL -d 1 -s 502 501 1440 one.i2i
addlines one.i2i 110215ligand_0_491_cell1_interpS.rpts - | maskedsmul -O 0 one.i2i - cell1_binary.i2i
# could do the same for a second object (etc), with create_dcimL -d 2, then use mathi2i
# to add the images before analysis
com_2d_objs cell1_binary.i2i > cell1_binary.stats
exo4
Phase 1: Associates each voxel in inimage.i2i which is > threshold with a maxima in the image. It finds
the maxima by doing a steepest ascent walk in the image. Writes summary info about regions found to
stdout in the same format as countobjs. Only the location of the maxima found in phase 1 are passed to phase 2.
All vesicles must start out as maxima. (see also -readmax option)
Phase 2: In each rolling window of time points (see -w), examines the intensity over time of each maxima
(found in previous step) to see if it
meets certain criteria (i.e., looks like a vesicle fusing with the plasma membrane - undergoing
exocytosis). If it matches this criteria it prints out the "path" of the maxima and its
pixel value and iod value. These paths can be converted to rpts format for
addlines or xmgr format
using found4.pl.
The fusion model: think of intensity as shown below. once the vesicle primes its intensity jumps. when it fuses
it drops back down.
peak intensity profile:
pintens ----------------
| |
| |
| |
tintens ------------- ------------------
0 t1 t2 t3
0 t1 |prise| |ffall|
t1: time of tethering (should stop moving)
t2: time of priming (peak intensity increases from tintens to pintens)
t3: time of fusing (peak intensity drops back down, but iod should stay high and possibly even increase a bit)
prise is the time it takes to rise froom tintens to pintens during priming (docking)
ffall is the time it takes to fall from pintens to tintens during fusion.
exo4 is similar to exo3 but has slightly different options and checks all possible fusion times
simultaneously. exo3 only picked the latest (in time) possible fusion event based only upon intensity
changes during the falltime. exo4 considers what is happening elsewhen in the window before deciding
on a fusion time. It picks the fusion candidate (iod rise and intensity fall) which is closest in time to
the maxima disappearing (-check_max) and which has a stationary period (-primedtime) within the
specified time range. Hopefully this makes it less likely to make mistakes.
Should work in 2d or 3d. Only reads in a window of time points at once
so should be able to handle large images.
Usage:
exo4 [options] inimage.i2i > inimage.count
Options:
options related to input:
-T startt stopt : only examine these time points (0-indexed, inclusive)
-p numskip numuse : patterned read. skip numskip z planes, then analyze numuse zplanes, then repeat.
This is useful if the acquisition protocol was, e.g., 1 fps for 1 min, 20 fps for 3 seconds
repeated over and over, and you only want to look at the 20 fps data.
numuse must be >= window size (see -w option). Pattern starts at t=0 even if -T is
specified (but startt time points are still skipped).
-w # : number of time pts in a window, default = 100
a negative number means read in entire dataset at once (one big window)
-jump # : jump # time pts before examining next window of time pts. This doesn't skip data, just
analyzes a "window" less often (so more has changed). Default = 0
which means the very next time pt is added to window (and oldest dropped) and then
the window of data is re-evaluated. May not work with -p option.
-r rptsfile : read in rptsfile. should have one closed object in it. Uses this as a mask for all the planes.
-d tdim : the time dimension of the image (over-rides anything stored in the header)
-b #: : blur (smooth) image before looking for objects (maxima), using a window of the specified
: halfwidth (in pixels) in x,y, and z (z width will be set to 0 if 2*width > zdim).
Phase 1: options related object finding via ascent (see objs_via_ascent2d), info printed out.
If an object doesn't satisfy these criteria, its maxima is not kept, so it is not considered a maxima
for the next stage of processing which tracks maxima into paths and checks if they are fusion events.
The iod and size specified here are also used later, but the next phase just looks with the square iod region.
-t threshold : any voxel > threshold in inimage.i2i is tracked to a 3d maxima (default = 0.0).
-i miniod maxiod : erase all objects whose iod doesn't fall in the specified range (default: 1-1000000000)
-s minsize maxsize : erase all objects whose size (in voxels, inclusive) doesn't fall in the specified range
(default = 1-1000000). note: object size and iod might be large since all voxels which
connect up via steepest ascent algorithm to the maximum count in this first phase.
-strict_max : ignore "maxima" which are really on a plateau (option might cause
saturated maxima to be missed)
-readmax file : skip phase 1, instead just reading maxima in from the specified file
Phase 1b: noptions related to fitting Gaussians (not used for anything yet):
-g # : fit Gaussians (+ dc term) to each object found (eventually I will use this for something).
only includes pixels in object within # pixels from the brightest point.
-G # : same as -g, but uses gradient info of fuction during fit (i.e., a different fit routine).
The -G version seems a bit buggy.
-n : normalize the Gaussian fom (shouldn't affect individual fits, just allow comparison between fits
Phase 2: options related to "paths" over time:
Maxima must exist for 0 time pts, be stationary for 0 time pts prior to disappearing
While a maxima is present, a pixel within range only counts as part of object
if above threshold (-t option). While a maxima is present,
a region will only be considered if size (pixels > thresh and within range) is within size (-s)
and iod is within iod (-i option).
-iodsize # check iod within +/- range pixels., default = 3
-maxspeed # max distance a maxima can move (in pixels) between time points, default = 5
-diststill # how far (in x or y) in pixels a maxima can move and still be considered "still",
(this is the location compared to last position of maxima) default = 0
-fall time frac maximum # of time pts peak intensity can take to show the specified fractional decrease
when vesicle is fusing. default = 10 and 0.300000
-iodrise frac the fractional increase in iod which must be seen
when vesicle is fusing (see -fall). default = 0
-check_max # vesicle must also change from max to not a max within # timepts after fusing
-minpeakval # peak intensity value must exceed this when vesicle is fusing (-fall time parameter)
-primedtime # # minimum and max # of time pts vesicle must be primed before fusing.
options related to output:
-debugpath x y z print out debug info about the path that passes near (within 5 pixels) here
(fgrep output for the word: debugpath). can be used 10 times. 1-indexed.
A -1 for z, means debug any path near x and y, regardless of z value (only if -L used).
-neardebug # for -debugpath, definition of near, in pixels, default = 5
-onlydebug : only print out info for debug paths. ignore all other paths.
-a : print all info about each object found (like countobjs does) rather than just summary stats
: for each time point.
-o outimage : create an ouput image of marked objects (object ids reused each time point). Max of each
: object will be marked with the negative of the object id.
-fit_model : fit a square wave to intensity profile of each vesicle found to be fusing (and to debugpaths)
not used for anything yet.
-v : verbose
-V : very verbose. may produce huge files. put this as first option to get option parsing info out.
Examples:
max2d_simple -t 50 inputimage.i2i -p max.pts
exo4 -v -r mask.rpts -d 600 -b 1 -w 100 -jump 20
-strict_max -readmax max.pts -s 10 1000 -i 10 1000000 -t 1
-duration 20 -timestill 20 -maxspeed 5 -iodsize 5 -diststill 2
-rise 25 .3 -fall 15 .3 -iodrise .1 -minpeakval 60 primedtime 30 90
-neardebug 2 -debugpath 152 161 237 -debugpath 134 124 -1
inputimage.i2i >& inputimage.out
graph_exo3.csh inputimage.out, which does the following:
findpaths_exo3.pl -trackxmgr=tmpavg.xmgr inputimage.out
findpaths_exo3.pl -trackxmgr=tmppeak.xmgr -peak inputimage.out
findpaths_exo3.pl -trackxmgr=tmpfit.xmgr -finalfit inputimage.out
findpaths_exo3.pl -trackxmgr=tmpdist.xmgr -distance inputimage.out
xmgr -graph 0 tmppeak.xmgr tmpfit.xmgr -graph 1 tmpavg.xmgr -graph 2 tmpdist.xmgr &
in xmgr: Edit->Arrange Graphs, select all the graphs, then set rows to 3,
Edit-Preferences set Crosshair cursor, linked scrolling.
findpaths_exo3.pl -trackrpts=tmp.rpts -printfusion=4 inputimage.out
addlines inputimage.i2i tmp.rpts - | play -P -1 R -P -2 G -
dave -n 0 -u -K -I inputimage.i2i tmp.rpts
source code in /home/lml/krypton/facil
see also:
objs_via_ascent, ~lml/krypton/Huang/TIRF4/found4.pl, run7
#/home/lml/krypton/bin/exo4
exo6
This programs find vesicle fusion events in a 2D time series.
Each maxima (-readmax) gets a size and iod associated with it. Only pixels > threshold (-t)
within the iod region (-iodsize) count toward the iod and size. Once the maximum disappears
(-nomax_length) the region centered around the previous location of the maximum is used.
Maxima are tracked from one time pt to the next by finding the closest maxima (within -maxspeed)
to its previous location. Tracks continue after the maximum disappears for -nomax_length time pts.
At the end of this time, the path is then analyzed to see if it meets criteria for a fusing vesicle.
All time pts during which the maxima exists must have an iod within the correct range (-i) and
and correct size (-s). The fusion time is identified by the intensity of the maxima falling
fast enough and far enough (-fall); while within the same interval the iod rises enough (-iodrise).
Fusion time is then further refined to be the time of largest iod increase (between adjacent time pts)
within this interval.
A fusion event may also need to be the correct time interval from when the maxima disappears (-check_max).
It may also need to be significantly brighter than the background (-minpeakratio).
It may be required to have a minimum peak value during fusion (-minpeakval).
Once the fusion time is found, the program finds the stationary (-diststill) time interval prior to fusion.
This (priming) time interval must be within certain constraints (-primedtime) for the path to be considered
representing a valid fusion event.
exo6 is similar to exo4 but reads in one time point at a time, there is no concept of a "window"
It only works on a 2D time series
Usage:
exo6 [options] -readmax max.rpts inimage.i2i > inimage.count
Options:
options related to input:
-T startt stopt : only examine these time points (0-indexed, inclusive)
-r rptsfile : read in rptsfile. should have one closed object in it. Uses this as a mask for all the planes.
-d tdim : the time dimension of the image (over-rides anything stored in the header)
-b #: : blur (smooth) image before looking for objects (maxima), using a window of the specified
: halfwidth (in pixels) in x,y, and z (z width will be set to 0 if 2*width > zdim).
-readmax file : read maxima in from the specified file (this is REQUIRED)
options related to finding fusion events:
-t threshold : any voxel > threshold in inimage.i2i is tracked to a 3d maxima (default = 0.0).
-i miniod maxiod : erase all objects whose iod doesn't fall in the specified range (default: 1-1000000000)
-s minsize maxsize : erase all objects whose size (in voxels, inclusive) doesn't fall in the specified range
(default = 1-1000000). note: object size and iod might be large since all voxels which
connect up via steepest ascent algorithm to the maximum count in this first phase.
-iodsize # check iod within +/- range pixels., default = 3
-maxspeed # max distance a maxima can move (in pixels) between time points, default = 5
-diststill # how far (in x or y) in pixels a maxima can move and still be considered "still",
(this is the location compared to last position of maxima) default = 0
-fall time frac maximum # of time pts peak intensity can take to show the specified fractional decrease
when vesicle is fusing. default = 10 and 0.300000
-startfall time how close (in timepts) in time the peak intensity must start to fall in relation
to the fusion time (biggest jump in iod between two consecutive time pts). default = 1
-iodrise frac the fractional increase in iod which must be seen
when vesicle is fusing (see -fall). default = 0
-check_max min# max# vesicle must also change from max to not a max >= min# timepts and <= max# timepts after fusing
-nomax_length # only analyze a path # time pts after max first disappears
the default (which is also the minimum allowed value) = 1
-minpeakval # peak intensity value must exceed this sometime while vesicle is fusing (-fall time parameter)
default value = 0
-minpeakratio ratio# time# dist#: the ratio of maxval/(median bkgnd value) must >= ratio# within
time# time steps of fusion.
bkgnd values are those pixels whose distance from max is >= dist# and <= iodsize
defaults: not to check this at all
-primedtime # # minimum and max # of time pts vesicle must be primed before fusing.
options related to speed:
-nomedian if -minpeakratio is defined, this causes the avg bkgnd value to be used instead
of the median value (calculation of median can be expensive).
-buffered_read # read # z planes at a time to speed up disk I/O. Default is 1 z plane at a time
options related to output:
-debugpath x y z print out debug info about the path that passes near (within 5 pixels) here
(fgrep output for the word: debugpath). can be used 10 times. 1-indexed.
A -1 for z, means debug any path near x and y, regardless of z value (only if -L used).
-neardebug # for -debugpath, definition of near, in pixels, default = 5
-onlydebug : only print out info for debug paths. ignore all other paths.
-a : print all info about each object found (like countobjs does) rather than just summary stats
: for each time point.
-o outimage : create an ouput image of marked objects (object ids reused each time point). Max of each
: object will be marked with the negative of the object id.
-v : verbose
-V : very verbose. may produce huge files. put this as first option to get option parsing info out.
Examples:
max2d_simple -t 50 inputimage.i2i -p max.pts
exo6 -v -r mask.rpts -d 600
-readmax max.pts -s 10 1000 -i 10 1000000 -t 1
-timestill 20 -maxspeed 5 -iodsize 5 -diststill 2
-fall 15 .3 -iodrise .1 -minpeakval 60 primedtime 30 90
-neardebug 2 -debugpath 152 161 237 -debugpath 134 124 -1
inputimage.i2i >& inputimage.out
graph_exo3.csh inputimage.out, which does the following:
findpaths_exo3.pl -trackxmgr=tmpavg.xmgr inputimage.out
findpaths_exo3.pl -trackxmgr=tmppeak.xmgr -peak inputimage.out
findpaths_exo3.pl -trackxmgr=tmpfit.xmgr -finalfit inputimage.out
findpaths_exo3.pl -trackxmgr=tmpdist.xmgr -distance inputimage.out
xmgr -graph 0 tmppeak.xmgr tmpfit.xmgr -graph 1 tmpavg.xmgr -graph 2 tmpdist.xmgr &
in xmgr: Edit->Arrange Graphs, select all the graphs, then set rows to 3,
Edit-Preferences set Crosshair cursor, linked scrolling.
findpaths_exo5.pl -rptsfile=tmp.rpts -rptstracks -printfusion=4 inputimage.out
addlines inputimage.i2i tmp.rpts - | play -P -1 R -P -2 G -
dave -n 0 -u -K -I inputimage.i2i tmp.rpts
source code in /home/lml/krypton/facil
see also:
objs_via_ascent, track_back2, ~lml/krypton/Huang/TIRF4/findpaths_exo5.pl, run7, graph_exo3.pl
#/home/lml/krypton/bin/exo6
find_2d_objs
For each z slice (time point) in the image, find 8-connected objects. Print info to stdout.
Only voxels > threshold can be part of an object (default = 0.000000). Doesn't use much memory (reads in a slice at a time).
Should handle our regular (short int) image format and floating point images.
Usage:
find_2d_objs [options] image1.i2i
Options:
-s min max: object must be within this size in pixels (inclusive), default = 1 100000
-b min max: object IOD must be within this range (inclusive), default = 1.000000 1000000000.000000
-t #: threshold for image1.i2i default = 0.000000
-T #: adaptive threshold. # = pixel intensity/mean intensity for each time pt. must be > # to be in object.
If -o specified, only pixels under the mask counted calculating mean intensity.
-i thresh.i2i: use with -T option if desired. thresh.i2i is used to calculate the threshold intead of image1.i2i
If -o specified, then that mask will also be applied to thresh.i2i.
-n image.i2i: write out new image with all voxels not part of an object set to 0
-z low high: only analyze z slices from low to high (1-indexed, inclusive).
-o mask.i2i: apply mask image to before analyzing it. only keep pixels in image1.i2i under mask pixels > 0.
mask.i2i must be in our standard image format (short int). If 3d, only first 2d image used.
-v: verbose
See also: pt_distances, max2d, max3dL, maxima, maxL, maxzL, float2intL, int2float, playexact
track, max2d_simple, countobjs
Example:
# find all objects between 5 and 30 pixels, minimum iod of 200, above a threshold of 100, write out new image:
find_2d_objs -t 100 -s 5 30 -b 200 1e9 -n newimage.i2i image.i2i > image.objs
Source code in ~krypton/lml/facil
find_blobs
/home/lml/krypton/bin/find_blobs
insufficient number of arguments
This program pads an image in z, then calls "invariant" (with -F 10) to produce multiple resolutions
of the original image along with its 2nd derivative in the gradient direction.
The blurred originals are used to find the location of the maxima. The derivative images should
have a zero crossing at the inflection points (eg, "edges"). The maxima should be in regions with
negative second derivative. This program then calls flood2zero to find the spatial extent of each
"blob" (bright region surrounding the maxima). Ideally this should just be a flood fill until
the zero crossing is found, but noise, etc. means that a lower threshold works better.
USAGE: find_blobs [options] inputimage.i2i outputroot
output extensions of files produced (in same directory as inputimage.i2i):
.ppts padded points coords (location of maxima in blurred, but not deriv, image).
.pts unpadded points, with z value transformed back to unpadded coordinates
.bpts unpadded points after filtering to keep bright points (\>= min_intensity)
.dpts bpts with "unordered" -> "ordered" and "break" -> "#moveto", for display in DAVE
_bres#.i2i blurred, but no derivatives, at resolution level = # (bigger # = lower res.)
_dres#.i2i blurred & derivatives taken, at resolution level = #
_dres#_obj.i2i objects found in *_dres#.i2i by floodfilling from points in *.bpts
_dres#.obj statistics on *_dres#_obj.i2i
_flood.out output from flood2zero
.out output from invariant
options:
-m #: any maxima less than this will get filtered out, default = 1
-d #: any derivative voxel with a value >= # will stop the flood fill
this is in the original, floating point coord system (prior to
rescaling to write out deriv. image). default = -5
-p #: pad data in z so that we don't have wrap around problems, default = 10
-z #: number of z planes (in original image) to skip in front and back when
finding blob voxels - the derivative calculation tends to create negatives
in the first and last few planes. default = 2
-r #: number of resolutions to calculate, default = 1
-e #: maximum spatial extent of object in pixels (ie, radius), default = 5
this is needed since the floodfill can otherwise escape and fill huge
volumes (the first and last z slices tend to be all negative, along with
negative values in the background regions).
-s #: maximum number of voxels in object, default = 300
-N #: normalize image prior to use so its max = #, default = 10000
-b sigx sigy sigz: standard deviation,in pixels, of Gaussian used in blurring
(passed as args for -s option of invariant), default = 2 2 1
examples:
find_blobs control3cluster4r_r_cell.i2i c3c4F10
dave -n 0 -u -z 1 -I control3cluster4r_r_cell.i2i -I c3c4F10_dres0_obj.i2i -S 2 1 0 c3c4F10.dpts
source for this cshell script is in /storage/big1/lml/was_invitro/unc/curvature
find_vessels
This program takes an image and scans it vertically in y at dx intervals. It counts the number of times a vessel of at least the specified width is entered. A vessel is any pixel with the specified intensity value. Each z slice in analyzed separately (and read in sequentially). A count of vessels vs time and dx displacement is written to stdout. See /home/lml/krypton/Corvera/grant2010. Usage: find_vessels [options] image.i2i options: -z start stop: zplanes to analyze. 0-indexed inclusive. default is all z planes -i #: intensity of a pixel in the vessel. default = 32767 -w #: minimum vertical width of vessel in pixels, default = 2 -d #: dx step size, default = 100 -v: verbose Source in /home/lml/krypton/facil See Also: Examples:
fit3gaussians
# /home/lml/krypton/bin/fit3gaussians -h
This fits a 3 component (Gaussian diffusion) model to Joan Politz's data.
Usage:
fit3gaussians [options]
Description:
Fits a three component Gaussian model to time series data.
Options:
-a : rather than having data fit a 3D diffusion model, have it fit a 2D model.
-d # # : only look at data sets from # to # (inclusive, first data set = 0).
timings for the available data sets are (in seconds after uncaging):
before_uncaging 0.050000 0.550000 1.050000 1.550000 11.550000 21.549999
31.549999 0.000000 0.000000 0.000000 0.000000 0.000000
0.000000
default data sets to look at are 1 to 4
-e # # : change the time in dataset # (0-indexed) to # (in seconds)
-f file : file name for test data (as produced by "distances" program).
-F file : file name for times for all the datasets from file, one time per line.
this also changes the default range to be all the sets
(use -d after this on the command line if you want to change the sets used).
note: there must be less than 1000 datasets.
-n # # : number of function parameters and variances used for
each parameter (default 10 paramters and 5 variances)
-p # # : set function parameter number to some value (first # can be 0 to
the maximum number of parameters)
-s # # : set function paramter # start value to some value (first # can be 0 to
the maximum number of parameters)
-S # # : set function paramter # variance value to some initial value (first # can be 0 to
the maximum number of parameters)
-r # # #: set function paramter # range values to some initial values (first # can be 0 to
the maximum number of parameters)
-t # : maximum number of random trials to search for parameters (default=100).
-T # : search terminates when # successive times the smallest variance search is best. (default=5)
-m # : maximum number of times to perform search, regardless of best variance (default=1000).
-M # : maximum number of times try (for each parameter) to find a point within allowed range
default = 100
-D func #: print debug info. for specified function at level #
func 1: ars, levels: 1-4 with 4 being most info.
func 2: radial search, levels: 1-2
func 3: finding a test point, levels: 1-2
-P : just print out the data and theoretical curves for the start point and terminate.
example (see ~lml/vision/Politz/run)
default settings are:
t1 in seconds : range: -4.900E-02 to 5.500E+00, start: -2.500E-02
D1 in microns^2/sec : range: 1.000E-02 to 1.000E+02, start: 5.000E+00
mag1 in intensity/micron^3 : range: 5.000E+01 to 3.000E+04, start: 3.000E+03
dc value : range: 0.000E+00 to 2.000E+03, start: 2.000E+02
D2 : range: 1.000E-01 to 1.000E+03, start: 1.000E+02
mag2 : range: 5.000E+00 to 3.000E+04, start: 3.000E+02
t2 : range: -4.900E-02 to 5.000E+03, start: 5.000E+02
D3 : range: 1.000E-01 to 1.000E+03, start: 1.000E+02
mag3 : range: 5.000E+00 to 3.000E+04, start: 3.000E+02
t3 : range: -4.900E-02 to 5.000E+03, start: 5.000E+02
flood2zero
This program takes an original 3D image, a 2nd derivative image
as produced by invariant -F 9 -b, and a list of 3d maxima and produces
a new image which is just those voxels from orig.i2i that are negative
(the 2nd derivative of a bright spot is < 0) in the derivative image
and connected to a 3D maxima. So it is used in conjuction with invariant
to identifiy bright blobs (eg, synapses) in a 3D image.
Usage: flood2zero [options] orig.i2i deriv.i2i image.pts newimage.i2i
options:
-b # : which resolution of maxima to use (object=noderiv_maxima# in pts file)
-v # : instead of using 0 as the object boundary cutoff, use #
-z low hi: zrange to examine, zero-indexed, inclusive.
-d dist: max chessboard distance (in voxels) from each point to flood fill.
-s size: maximum size in voxels of an object (will truncate to this size), default= 10000
-o file: save object (blob) statistics to specified file. This is very similar to
what countobjs would produce when applied to newimage.i2i except newimage.i2i
can have blobs which touch (due to the size contraints here) and hence show
up as one large blob instead of two smaller blobs. Also, if the header for the
image has a scale factor in it (which invariant writes) this scale factor will
be used to convert all statistics back into original unscaled values.
note: entire image name (with extension) is NOT required.
note: a - in place of an image name means stdin or stdout.
note: a compressed image will be automatically be uncompressed.
[source in /usr/people/lml/invitro/unc/curvature/flood2zero.c]
gsearch
program to do graph search on a slice of edge image USAGE: example gsearch -d name -x # -y # -a # -b # edge.i2i text.gpts edge.i2i : magnitude input image text.gpts: output is a graph points (gpts) text file having the same format as rpts file new boundary is stored under z=slice # (default: 1) Options: -d option specifies name of image containing direction of each edge pixel of edge.i2i -k option specifies constant of proportionality to be used with filgra (default: 1.0) -x start x coordinate. -y start y coordinate. -a stop x coordinate. -b stop y coordinate. -h prints this message. Source code in /storage/big1/lml/jac/was_invitro/graph
histogram2
Usage:
histogram2 [options] boundary.i2i aimage.i2i bimage.i2i
Description:
This program creates a number of histograms of intensity or volume as a function of
distance of the object voxels from the nearest background voxel.
Requires:
boundary.i2i - boundary image created with boundary or createBoundary.
object voxels are those with a value of 6
background voxels are those with a value of 4
border voxels are those with a value of 2
all other voxels are ignored.
NOTE: by default this is assumed to be padded by 7 pixels all around.
ie, IT IS LARGER THAN aimage.i2i and bimage.i2i
createBoundary does this. see -p if you don't want this default.
aimage.i2i - data image a.
bimage.i2i - data image b.
Returns:
aimage_H.histo - output file with histogram info (-H can change this).
Output file format (10 columns):
1. bin number (i.e., distance from background) (each bin = 0.1 pixels)
note:a pixel touching the boundary is a distance of .5 (center is .5 from edge)
2. histogram of intensities in image a.
3. number of pixels in this bin (i.e., volume at this distance).
4. histogram of intensities in image b.
5. histogram of intensities in image a > athreshold (intens-thresh if -s option used).
6. histogram of intensities in image b > bthreshold (intens-thresh if -s option used).
7. histogram of intensities in image a, where values in a are such that
if (image a > athreshold) and (image b > bthreshold), a=a (a-athresh if -s option) else a=0.
8. histogram of intensities in image b, where values in b are such that
if (image a > athreshold) and (image b > bthreshold), b=b (b-bthresh if -s option) else b=0.
9. # of pixels in image a > image a threshold (at this distance), like 3 but only
counting voxels > threshold.
10.# of pixels in image b > image b threshold (at this distance), like 3 but only
counting voxels > threshold.
Input Options:
-o : border pixels are background voxels (default is object voxel)
-p x y z :padding in x,y,z included in the boundary image (default 7 in all directions).
(boundary image found in the programs boundary and
createBoundary are by default padded by 7 pixels
on all sides of the image.
Distance Options:
-P x y z : don't find histogram of all pixels to boundary, just the distance
of this pixel from the boundary. Can be used multiple times on command line.
(x,y,z) is 1-indexed (like play) aimage.i2i coordinate values.
-d #: maximum distance (in pixels) to check, default = .5*(min dim)
Any pixel further than the maximum distance is considered to be at the maximum distance.
This apply also for the -ta and -tb distance based thresholds.
-x # : set x weight to # (default 1.0).
-y # : set y weight to # (default 1.0).
-z # : set z weight to # (default 1.0).
-F : Fast histogram - voxel level (default fast).
-S : Sub-voxel resolution histogram(default fast).
-Y # #: limit Y direction from # to # (default off).
If the long axis of the cell is in the in the y direction,
and the cell is cut off, using the -Y option will limit searching
for the boundary in that direction, i.e. the ends past these values will
not be considered part of the boundary.
(similarly true for -X and -Z options)
-X # #: limit X direction from # to # (default off), inclusive, 1-indexed.
-Z # #: limit Z direction from # to # (default off).
-c: pixels outside the image are considered border pixels. For example, if the outlines
used to create boundary.i2i were in z=1, but you want pixels inside the z=1 boundary
to be counted as being 1 pixel away from the boundary (eg, from the phantom z=0 slice)
use this option.
Threshold Options:
-a # : set aimage threshold to # (default 0.000000).
-b # : set bimage threshold to # (default 0.000000).
-s : subtract thresholds (and then clamp to >=0) rather than simply keeping intensities > threshold.
This is more like what you might want to do to subtract off background or nonspecific flourescence.
-ta athresh.txt : apply different thresholds at different distances to aimage.i2i.
athresh.txt as produced by analyze_histo (applied to -f aimage_H.i2i output)
(i.e., 2 columns: bin # (tenths of a pixel) and threshold).
-tb bthresh.txt : like -ta, but for bimage.i2i
-ra # : rescale athresh.txt prior to use, multiply values in file by #.
-rb # : rescale bthresh.txt prior to use, multiply values in file by #.
Ouput Options:
-f aimage_H.i2i xdim ydim: print out a full histogram of aimage. This is a
2D image (eg, aimage_H.i2i) with the specified x and y dimensions.
The x direction is pixel intensity, and the y direction bin number (tenths of a pixel) and
the pixel value wll be set to the number of voxels at that distance and that intensity.
Specify -1 for xdim or ydim if you want that dimension calculated automatically
(xdim will be approx. max intensity in image and ydim about 10*max dist, see -d option).
Explicitly set the dimensions if you will want to add this image with others (since they must all
be the same dimensions), e.g., to average data from a set of control files.
note: -S option will cause the "number of voxels" to appear to change.
Use the analyze_histo program to produce a distance dependent threshold file from this (e.g. to read
back into histogram2 on another pass via the -ta or -tb option). This image can be large (25MB).
-H file: Output filename for histogram (instead of aimage_H.histo).
-A aimage_thresh.i2i : write out aimage.i2i after threshold is applied (for visualization purposes)
these are the pixel values used to produce column 5 of aimage_H.histo
-B bimage_thresh.i2i : write out bimage.i2i after threshold is applied (for visualization purposes)
these are the pixel values used to produce column 6 of aimage_H.histo
Miscellaneous Options:
-D distimage.i2i : print the distance image (for debugging). Each voxel is 100 times its distance
from the membrane (a pixel neighboring the background has a distance of .5 pixels).
This can also be used by mask_image to then mask desired pixels.
-h : print this message.
Notes:
All the images should be oriented in the same direction.
Entire image name (with extension) is required.
A - in place of an image name means stdin or stdout.
Example:
create_dcim -d 4 -s 234 294 24 -| addlines -P -1 - rob15_nucleus.rpts -|
flood -e -1 -E 2 -I 6 -b 4 - mask.i2i
histogram2 -p 0 0 0 -Z 2 23 -d 40 -a 3552 -b 7767 -z 1.34 mask.i2i rob15f_rtm.i2i rob15r_rtm.i2i
Example: use a variable distance threshold to subtract background flourescence from dual labeled images:
addlines -P -1 control_aimage.i2i control_membrane.rpts -| flood -e -1 -E 2 -I 6 -b 4 - mask.i2i
# create a full histogram so we can use a variable threshold based upon distance from membrane:
histogram2 -p 0 0 0 -d 40 -z 1.34 -f controla_H.i2i mask.i2i control_aimage.i2i control_aimage.i2i
# create the variable threshold file:
analyze_histo controla_H.i2i > controla_H.thresholds
# repeat for control bimage (e.g., second wavelength):
histogram2 -p 0 0 0 -d 40 -z 1.34 -f controlb_H.i2i mask.i2i control_bimage.i2i control_bimage.i2i
analyze_histo controlb_H.i2i > controlb_H.thresholds
# now apply these thresholds to the real dual labeled (not the control) data:
addlines -P -1 real_aimage.i2i real_membrane.rpts -| flood -e -1 -E 2 -I 6 -b 4 - real_mask.i2i
histogram2 -p 0 0 0 -d 40 -z 1.34 -s -ta controla_H.thresholds -tb controlb_H.thresholds \
real_mask.i2i real_aimage.i2i real_bimage.i2i
See Also:
boundary, createBoundary, analysis, analyze_histo, threshold, nucleus_overlaps, mask_image, Corecell.pl.
Source code in /storage/big1/lml/jac/was_invitro/Projects/boundary/boundary
image2ptsL
This program takes an image, and identifies boundary pixels of objects. Boundary pixels are foreground pixels which have at least one backgound pixel among their 8 neighbors. In addition, any foreground pixel which is on the edge of a 2D slice is also considered a boundary pixel. These boundary pixels are written out in "addlines" format. Each zslice is analyzed sequentially. Points are unordered, and they are all considered part of one object. Points in the pts file have their value set to -1 Usage: image2pts [options] image.i2i pointfile.pts options: -t #: threshold, above which is foreground. Default = 0 note: entire image name (with extension) is NOT required. note: a - in place of an image name means stdin or stdout. note: a compressed image will be automatically be uncompressed.
invariant
This program takes an image, blurs it with a Gaussian of
specified standard deviation and then takes derivatives of the image
It does all of this in the frequency domain.
outfile is the root name for derivative of blurred images (outfile_dres#.i2i, unless -M), plain blurred
images (-b option, outfile_bres#.i2i) and/or pts files (-m or -M options, outfile.pts).
Any function which is a ratio of sum of products of derivatives (eg, (LxxLy + LyzLx)/(LyLy+LzLz))
can be calculated. Edit file init_invariant.c in
/usr/people/lml/invitro/unc/curvature and recompile on molmed if you want a function which
is not listed here.
Usage: invariant [options] infile.i2i outfile
options:
-F #: function to calculate. Default = 1. Available functions are:
-F 1: LxLx+LyLy+LzLz : 3D gradient squared
-F 2: .5(Lxx+Lyy+Lzz)/sqrt(LxLx+LyLy+LzLz) : normalized 3D Laplacian
-F 3: LxLx+LyLy : 2D gradient squared
-F 4: sqrt(LxLx+LyLy) : 2D gradient
-F 5: 2(LxxLyy-Lxy^2)/(Lxx^2 + Lyy^2 + 2Lxy^2): 2D light or dark blobs (p. 252 IPMI '91)
-F 6: 2(LxxLyy-Lxy^2), same a -F 5, no normalization (so noise isn't magnified).
-F 7: mean curvature of 3D isointensity surface normalized by gradient squared
-F 8: mean curvature of 3D isointensity surface, no normalization.
-F 9: 2nd der. in the grad. direction times gradient squared.
-F 10: 2nd der. in the grad. direction (should = 0 at locations of edges)(use -u option).
-F 11 XYZ e.g., -F 11 021 take zero derivatives in X direction, 2 in Y, 1 in Z (Lyyz).
by calling invariant multiple times with various XYZ and using the -g option you
can create a bunch of derivative images which you can then read in and combine
however you like (e.g., my curvature_eigenvectors program)
-N #: normalize image intensities prior to use to be between 0 and #
this performs: newval = #*(val-minval)/(maxval-minval)
-u #: during division (-F 2,5,7,10) set divide by 0 to this integer, default=0.
note: this is the final output pixel value, after any scale factors (-r or -R).
a good value is -32768, the minimum possible output pixel value.
-s sigx sigy sigz: standard deviation in spatial domain of Gaussian.
-S sigx sigy sigz: standard deviation in frequency domain of Gaussian.
default = 3.402823E+38 3.402823E+38 3.402823E+38
-l # res2 res3 ... : # = number of different resolution levels to calculate.
res2 ... res# are the relative sigmas for each resolution level after
the first one. eg, -l 3 .9 .7 means that 3 blurred levels should
be calculated and the sigma (in the freq domain) of the second
level should be .9 of the first level (ie, more blurred), and the 3rd
level should be .7 of the first level (as specified with -S).
-L # factor: # = number of resolution levels. factor is the relative sigmas in
the spatial domain for each level relative to the previous level.
-e epsilon: cutoff for Gaussian intensity calculation.
default = 0.040000
-r: apply scalefactor to output so min= -32766 or max= 32766. This is useful
especially when all the values would otherwise be < 1.
This also prints to stderr the min and max values before rescaling.
-R #: rescale output by multiplying all values by #.
-m: calculate coords of 3D maxima after blurring and taking derivatives.
Creates an outfile.pts file.
-M: same as -m, but also prevents writing out blurred images.
-b: calculate coords of 3D maxima after blurring (but before derivatives). This
can be used with the -m option. Objects associated with this option will
be named "noderiv_maxima#", where # is the resolution number.
This is useful for passing to flood2zero to find the extent of the bright spots.
Also writes out blurred images (outfile_bres#.i2i).
-B: same as -b, but prevents writing out the blurred images.
-n #: maxima must have an intensity > # (must use -m, -M, or -b with this)
-f #: max pixel must be > all neighbors by at least # to be a maxima. default = 0.000100
-g: output a floating point image (no rescaling done). This output can be used
by my curvature_eigenvectors program. -u option still applies.
note: image dimensions which factor to 2, 3, or 5 go fastest with the FFT.
note: the entire image name is NOT required.
note: compressed images will be uncompressed.
note: a - in place of an image name means stdin or stdout.
see also: max3d, multi_gauss_deriv, flood2zero, find_blobs, blur_spatial, canny, zerosu.
source in /storage/big1/lml/was_invitro/unc/curvature
/home/lml/krypton/bin/invariant
linux_epr_x86_64-7-12-2012
epr UMMC/BIG Jul 12 2012
NAME
epr - Exhaustive Photon Replacement (EPR) restores contrast by removing
residual out-of-focus light and improves resolution while maintaining
numerical accuracy of 3-D images of specimens obtained with serial
optical sectioning from wide-field or confocal light microscopy.
SYNOPSIS
epr [options] before-image[.tif] after-image[.tif]
DESCRIPTION
epr performs regularized, iterative image restoration with a non-
negativity constraint. The before-image is a three dimensional (3-D)
TIFF image composed of rectangular, regularly spaced, optical sections.
Large images are decomposed into smaller, overlapping (in x and y only)
image segments for restoration, and the restored segments are recomposed.
The after-image is the restored 3-D image. All options must appear
before the image file names, but the order of options is not important.
epr has the following options:
-h Prints basic help on syntax and available options.
Required Parameters
-psf file[.tif] The 3-D point spread function image file. The psf must
match the optical configuration used for the before-
image. The psf depth (number of z-planes) need only be
less-than or equal-to twice that of the before-image.
Extra z-planes are symmetrically discarded.
-smoothness alpha The alpha value where alpha is typically 0 < alpha < RNL.
-sm alpha RNL is the residuals noise limit as reported by prepdata
(see also). Smaller values of a correspond to less
smoothing. (RNL)^2 is usually a good starting choice for
alpha.
-iterations n Maximum number of iterations to perform. Iterating may
-it n terminate earlier if convergence is detected. (see
-convergence)
Control Parameters
-scaling n where n is after image scaling factor. Used to prevent
-sc n integer overflow when saving after image to file.
Default=1.0
-convergence n Criteria for terminating iteration, where n is << 1.
-co n 0.00001 represents true convergence. 0.001 usually
achieves 90-95% convergence in about half the number
of iterations. Values of n larger than 0.001 are not
generally recommended. Default=0.001
-axial n1 n2 The z-axis object extrapolation beyond the sectioned
-ax n1 n2 image data where n1 is before first plane and n2 is
after last plane. n is in planes. By default,
extrapolation extends +-1/2 the z-axis extent of the point
spread function image and should be sufficient. Smaller
values of n1 or n2 have the effect of spatially
constraining the restored object and should be applied
carefully.
-time n1 The timepoint where you want the restoration to occur (0 indexed)
-t n1
-threads n The number of threads FFTW should use. Default=1
-th n
-transverse n The x-axis and y-axis object extrapolation in pixels.
-tr n The epr process extrapolates the restoration beyond the
bounds of each image segment in order to account for
exterior out-of-focus contributions. Default = 1/4 of X or Y
dimsenion PSF width, which ever is larger.
EXAMPLES
To see the command line syntax
% epr
or
% epr -h
Given specimen image mycell_.i2i and point spread function image mypsf_.i2i
let smoothness equal 0.0005, maximum number of iteration equal 250
and convergence equal 0.001. Perform single-resoltion(default) EPR:
% epr -psf mypsf_ -smoothness 0.0005 -iterations 250 mycell_ mycell_r
FAQ (Frequently Asked Questions)
see http://...
FILES
xcomp:/home/epr/epr
xcomp:/home/epr/cs_epr.lo
SEE ALSO
prepdata
preppsf
many_overlapsL
insufficient number of arguments This shell script calls overlap many times for a range of thresholds in both images. Unknown options are passed to overlap. Output is put into the specified output file This should then be processed by the program: create_images Note: a temporary file is created in the current directory. See also: create_images, overlap, probcolocal USAGE: many_overlaps [options] image1.i2i image2.i2i output options: -t low hi step :range of thresholds for image1.i2i default = 1, 101, 10 -T low hi step :range of thresholds for image2.i2i default = 1, 101, 10
map_to_surface
error:
There should be 3 or 5 main arguments (exluding options).
Arguments seen:
This program takes a surface (which are those voxels which are nonzero)
and one (or two) data images. It then maps (moves) the data voxels (nonzero
voxels) to the position of the nearest surface voxel. When done, one (or two) new
image is produced which has the data values at these new positions.
In addition, some statistics about the mapping are printed to stderr.
NOTE:A surface can be generated by Simple_2D_wrap or shrink_wrap. This "mesh"
representation of the surface (which DAVE can display using the -i option)
can be converted into a surface .i2i image for map_to_surface using
/usr/people/jac/cell/bin/make_surface -I surface.i2i -c 1 mesh.i2i surface.rpts
The resulting surface is made up of voxels. Voxels which are just below the
surface will be considered to be a distance of 1 voxel from the surface
(the distance between the centers of the two voxels).
Usage:
map_to_surface [options] surface.i2i image.i2i out_image.i2i [image1.i2i out_image1.i2i]
Options:
a second input image and output image can be specified on the command line.
-c: rather than mapping data values to the nearest surface voxel, map
a count of the number of data values which mapped to the
surface voxel. I.e., if 2 data voxels mapped to a surface voxel, a
2 is stored there (rather than the max of the 2 data values-the default).
-b: rather than mapping data values to the nearest surface voxel, or
a count, just map a 1 (ie, binary) onto the surface where ever a data
voxel maps (regardless of how many map there or their intensity).
-m: surface.i2i is actually a mesh file, not an image file
produced from a mesh file. In this case, only map data
values to surface vertices (since map_to_surface doesn't
yet know how to convert mesh files to complete surfaces
like make_surface).
-d #: minimum distance (in voxels) which data must be from the surface and
still be mapped onto it. Data closer than this is ignored.
By default all data is examined (ie, min_distance = 0)
-D #: maximum distance (in voxels) which data can be from the surface and
still be mapped onto it. Data further than this is ignored.
By default all data is examined (ie, max_distance = 1/2 the minimum
image dimension, this should include all of the data).
-x: exclude data voxels which are already on the surface (they will be zeroed).
(note: setting the minimum distance > 0 will not achieve the same effect since
that would still allow a data voxel on the surface to match to a surface
voxel at a neighboring position).
Source code in /storage/big1/lml/was_invitro/Moore/pkc
min_cylinder
MIN_CYLINDER program - Version 1.0 This program takes an initial cylindrical mesh surface and allows it to deform to minimize a function of surface curvature and stretch and image intensity Usage: min_cylinder [options] insurface infile outsurface OPTIONS CONTROLLING TERMINATION CRITERIA: [-n #] Maximum iterations [-c #] Minimum pixel changes per iteration OPTIONS CONTROLLING SURFACE SHAPE: [-w # # # # # #] Weights for Su Sv Suu Suv Svv E default = 0.20, 0.20, 0.10, 0.20, 0.10, -0.10 NOTE: for a 2D time series, the u direction is usually in the xy plane and v is the time direction. -b: use bilinear interpolation along the u coordinate direction to calculate the energy component due to the data. This option only works in conjunction with the -D option. This causes all data along the boundary in each xy plane to contribute to the energy of the surface, instead of just voxels at the locations of the vertices of the surface. NOTE: does not use weights specified by -W option yet. -a: same as -b but the average energy along the boundary is calculated instead of total, so long lines are not preferred. -p: same as -a but the average energy along each boundary segment is weighted by the percentage of total boundary length which that segment contributes. So long bad lines count more than short bad lines. -g: weight the data along the boundary by the dot product with the unit length vector normal to the gradient. Thus boundary segments which are parallel to the data gradient direction will count less than they otherwise would. Use with -b, -a, or -p option. -z #: the cost of a zero length line (before applying w6) default = 0, only valid with -a or -b option. -N: use "normal" curvature instead of parameterization dependent curvature calculation. This option should produce a more correct curvature calculation, so the program does not get fooled into thinking that small objects have smaller curvature. OPTIONS CONTROLLING ALLOWABLE SURFACE MOVEMENT: [-D] Restrict movement to 2 dimensions (x & y) [-s] Skip first and last planes of cylinder (so they won't move at all). [-S filename] filename of z planes to skip (do not use with -s). [-f] Fix first and last planes of cylinder to move only in x&y [-F filename] filename of z planes to fix to move only in x&y (do not use with the -f option). [-A] when the -F or -S flag specified, normally if two adjacent slices are specified in the rpts file then the surfaces on either side of these become independent of the each other. If -A is specified then they will still affect each other. OPTIONS CONTROLLING INPUT, OUTPUT, AND DEBUGGING INFO: [-W # # #] Weights for scaling x y z dimensions. [-r rescale] rescale image data from 0 to rescale before using. default = 1000.000000 If rescale <0 then no rescaling will be done. [-u] expect input image to be i2i - output also i2i. [-I #] write out vertex coords every # images written in unc image format. can be read in and displayed by wire. If SAVE_ENERGY is defined then energy of each vertex also saved. Currently SAVE_ENERGY is defined. [-i] Copy information fields flag [-t "title"] New image title [-d #] Debugging level, 1 (least) to 2 (most) See also: Simple_2D_Wrap, 2D_wrap, shrink_wrap
morph3d
This program does morphological processing on greyscale images. Currently the
default intensity it filters with is a constant, although different shapes and
intensity profiles are allowed. Think of a 2d image as a 3d binary array of voxels
where the height dimension is the intensity of the image at this x,y position.
All those pixels which are less than or equal to the height are 1, others are zero.
This object (made up of voxels which are 1) is the UMBRA of the image.
Dilation: Take the umbra of
the image and the umbra of the filter. Then the image dilated by the filter is
those voxels which get "reached" by the filter voxels when the filter umbra
is translated through the image umbra (i.e., move the origin of the filter umbra
everywhere inside the image umbra, all those voxels ever reached by the filter are
part of the dilated umbra).
For a constant intensity filter this reduces to just taking the
maximum of the image intensities at the filter's locations.
Erosion: similar to dilation, but now as you move the filter around, only keep
those voxels which, when the filter umbra is centered on them, permit all the
filter voxels to lie on top of image voxels (i.e., the filter umbra fits inside
the image umbra when centered on that voxel).Erosion is dilation of the background.
This reduces to a minimum for a constant intensity filter.
Opening: erosion followed by dilation, can be visualized by taking the filter umbra
surface and pushing it hard up under the image umbra surface. The highest voxels
reached by the filter surface represent the new filtered image surface.
(e.g. the "solvent or probe accessible surface" from below.
Closing: dilation followed by erosion. Dilation can be visualized as
flipping the filter umbra and rolling it along the top (outside) of
the image umbra and recording the lowest voxels reached. (e.g.,the solvent
accessible surface from above.).
If a 3D image is input, and a 3D filter shape is specified, then all analysis
is done using 3D morphological filtering. If a 2D filter is specified, then each
zplane is analyzed separately.
Usage: morph3d [options] inimage.i2i outimage.i2i
options:
-v: verbose, prints out which z slice is being analyzed to stderr.
-b #: any pixels <= # are background, ignore them. default = -32766
-h #: the maximum height (intensity) of the filter center relative to other
entries in the filter. The default =0, ie all entries in the
filter are at the same height. If a nonzero value is specified
a linear falloff in height to zero at the edges of the filter is
assumed.
-m: when -m is specified the filter's intensities must fit under the
image intensities without any intensities going negative. If this
not the case, then a zero value is put in that center pixel location.
For example, if we specify a filter which is 100 brighter in the
center than at the edge, and the filter is over a region that is 50
brighter in the center, then (if -m is not specified) that pixel value
might stay 50 (since the template can fit under the image intensity by
lowering itself so its edges have negative values). If -m
is specified, the filter will not fit and the pixel will get set to 0.
-f #: morphological filter shape. 6 = jack (ie, 3D cross, default)
7= 3D asterisk, 4 = 2D plus
1= 2D filled square, 8 = 3D cube, 2= 2D filled circle,
3= 2D asterisk, 5= 2D cross.
2D shapes are done one zslice at a time.
-l #: just use a single LINE, of specified orientation .
1 = LL-UR, 2 = UL-LR, 3 = Left-Right, 4 = Up-Down.
5 = In-Out in z, 6 = ULIn-LROut (a diagonal in 3D).
-s # # #: size of x,y, and z "radius" in pixels (4 4 2 default).
[-e|-d|-o|-c]: erosion (min), dilation (max), opening (e then d), or
closing (d then e). default is closing.
note: entire image name is NOT required.
note: a - in place of an image name means stdin or stdout.
note: the origin of the filter element is at its center.
see also: morph - this is an older, one z-slice at a time version.
multi_gauss_deriv
This program takes an image, blurs it with a Gaussian of specified standard deviation and then takes derivatives of the image It does all of this in the frequency domain. The FFT will be done in parallel unless the environment variable NPROCS is set to 1. FFT_MAXMEM variable defines the maximum memory the FFT will use. outfile is the root name for images and/or pts (-m or -M) files. Usage: multi_gauss_deriv [options] infile.i2i outfile options: -x order: number of derivatives in the x direction. default = 0 -y order: number of derivatives in the y direction. default = 0 -z order: number of derivatives in the z direction. default = 0 -s sigx sigy sigz: standard deviation in spatial domain of Gaussian. -S sigx sigy sigz: standard deviation in frequency domain of Gaussian. default = 3.402823E+38 3.402823E+38 3.402823E+38 -l # res2 res3 ... : # = number of different resolution levels to calculate. res2 ... res# are the relative sigmas for each resolution level after the first one. eg, -l 3 .9 .7 means that 3 blurred levels should be calculated and the sigma (in the freq domain) of the second level should be .9 of the first level (ie, more blurred), and the 3rd level should be .7 of the first level (as specified with -S). -L # factor: # = number of resolution levels. factor is the relative sigmas in the spatial domain for each level relative to the previous level. -e epsilon: cutoff for Gaussian intensity calculation. default = 0.040000 -r: apply scalefactor to output so min= -32766 or max= 32766. This is useful especially when all the values would otherwise be < 1. This also prints to stderr the min and max values before rescaling. -R #: rescale output by multiplying all values by #. -m: calculate coords of 3D maxima after blurring. -M: same as -m, but also prevents writing out blurred images. -n #: maxima must have an intensity > # (must use -m or -M with this) -f #: max pixel must be > all neighbors by at least # to be a maxima. default = 0.000100 note: image dimensions which factor to 2, 3, or 5 go fastest with the FFT. note: the entire image name is NOT required. note: compressed images will be uncompressed. note: a - in place of an image name means stdin or stdout. see also: max3d, gauss_deriv_unc, blur_spatial, canny, zerosu.
objs_via_ascent
Associates each voxel in inimage.i2i which is > threshold with a 3d maxima in the image. It finds the 3d maxima by doing a steepest ascent walk in the image. Writes info about regions found to stdout in the same format as countobjs. Writes an image with each pixel labeled with its region id to outimage.i2i. Usage: objs_via_ascent [options] inimage.i2i outimage.i2i > inimage.count Options: -t threshold:any voxel > threshold in inimage.i2i is tracked to a 3d maxima (default = 0.000000). -i miniod maxiod : erase all objects whose iod doesn't fall in the specified range -s minsize maxsize : erase all objects whose size (in voxels, inclusive) doesn't fall in the specified range Examples: objs_via_ascent -t 100 in.i2i in_id.i2i > in.count objs2dist.pl -dz=3 -jack=in_max.rpts -rpts=in_bb.rpts in.count in.count > in.comdist (don't really care about comdist here, just want rpts files) colorize_image -c in_id.i2i in.i2i inr.i2i ing.i2i inb.i2i (autoscales so max will be 255) dave -S all 1 0 -nocomp -I ing.i2i -I inr.i2i -Iinb.i2i -n 0 -u in_max.rpts in_bb.rpts (then turn blue channel on in dave) objs_via_ascent -t 100 in1.i2i in1_id.i2i > in1.count objs_via_ascent -t 100 in2.i2i in2_id.i2i > in2.count objs2dist.pl in1.count in2.count > in1and2.comdist closest_voxel -Q -t 0 0 -p -r 1 -w 1 1 3 in1_id.i2i in2_id.i2i > in1and2.patchdist /home/lml/krypton/Zhuge/BKandRyr/analysis3/graph.pl -showmatch in1and2.patchdist > in1and2_match.rpts source code in /home/lml/krypton/facil see also: countobjs countobjsmax3d objs2dist.pl colorize_image closest_voxel ~lml/krypton/Zhuge/BKandRyr/analysis3/run.pl channel_current ~lml/krypton/Zhuge/BKandRyr/graph.pl
overlap3
******* /home/lml/krypton/bin/overlap3
This program calculates the overlap (colocalization)
of two images. It is very similar to the overlap
program, but produces a whole set of numbers with
colocalization for each of many OFFSET DIRECTIONS at which
voxels are considered to be "in the same place"
In other words, suppose
two data voxels are not exactly at the same location in the 2
images, but are offset by 1 voxel in a specific direction. This
program would count that
as an overlap for, say, distance dx = -1, dy = 0, dz = 1.
Output is text written to stdout. The OFFSET displacement is the displacement for
image2. so OFFSET (-1,2,0) means image2 should be translated -1 in x (to the left), +2 in
y (up 2) and not at all in z to align it with image1, e.g., tranima -T -1 2 0 image2.i2i image2T.i2i.
Usage: overlap3 [options] image1.i2i image2.i2i [maskimage.i2i]
options:
-m maxdist: maximum translational distance that should
be checked (default = 1)
-x maxdist: maximum x translational distance that should
be checked (default = 1)
-y maxdist: maximum y translational distance that should
be checked (default = 1)
-z maxdist: maximum z translational distance that should
be checked (default = 1)
-s step : step size for translations
(default = 1)
-t thresh1 thresh2: only voxels in image1 > thresh1
and voxels in image2 > thresh2 are
considered for the overlap calculation.
default = 0 and 0.
-l: keep things less than or equal to the threshold in image1
(default is greater than the threshold)
-L: keep things less than or equal to the threshold in image2
(default is greater than the threshold)
-v threshold: all values greater than this value
are part of the mask (default 0)
-V maskvalue: this value is part of the mask
can be used up to 100 times
note: entire image name is NOT required.
note: a - in place of an image name means stdin or stdout.
source code in /storage/big1/lml/jac/was_invitro/Projects/overlap3
prune
This program takes an input image and prunes "dangling" paths from the image. The input image is assumed to have already been thinned to a 1 pixel-wide center line using thin3Ds. The output image can be converted to a pts file via im2pts. Which paths get pruned depends upon the options specified. By default, every end point is found and the path from the endpoint to first intersection point is deleted. The idea behind this program is to get rid of extraneous parts of the path produced by thin3Ds, to aid visualization. Usage: prune [options] image1.i2i image2.i2i options: -l #: only delete paths which are shorter than # pixels in length. -s x y z: x,y,z coords of a single path to keep. -S x y z: x,y,z coords of other end of the path to keep. (this should be used with -s). -d datafile: a file with lists of end points (start and stop) of paths to keep. Each line has a start and stop pt: (x1,y1,z1) (x2,y2,z2) -X lx hix: keep all paths whose endpts are within this x range (inclusive) If you specify any of X,Y,or Z, you must specify all 3. -Y ly hiy: keep all paths whose endpts are within this y range -Z lz hiz: keep all paths whose endpts are within this z range these 3 options are ANDed together. note: all coords are zero indexed (ie, first pixel is numbered 0, not 1) note: entire image name (with extension) is NOT required. note: a - in place of an image name means stdin or stdout. note: a compressed image will be automatically be uncompressed. /home/lml/krypton/bin/prune
shrink_wrap
/home/lml/krypton/bin/shrink_wrap
Usage:
shrink_wrap -R input.rpts input_image output_root [options]
input_image Image file for blurring - WITHOUT the .i2i ending
output_root Root name of output file (no .tail ending). will produce output_root.rpts
Options:
-B use sphere as initial surface (default cylinder)
-P name of file contaning point spread function
By default is a gausian psf
-r set blur3d to write output image as real
-R name of file containing rpts file
Must be used in the command line
-M name of file containing mesh
-U file: file is a pre-blurred attractant image in i2i image format
so input_image is not used for anything except
for image dimensions. see OUTPUT below.
-S Value for scaling blurred image
By default equals 1.0
-o value of overlapping in pixels along X and Y axes
By default equals 10.
-p value of zero padding in pixels along X and Y axes
By default equals 10.
-z value of zero padding in pixels along Z
By default equals 10.
-a memory allocation coefficient
By default equals
-w # # # # # #: Weights for Su Sv Suu Suv Svv E
u are pts in the plane and v are points in z or time
By default .4 .4 .2 .2 .2 -1.0
-W # # # Weights for x y z
By default 1.0 1.0 1.0
-D for two dimentional images in a time series
-d # dilate rpts file slices by #
By default 0
-F do not allow slices from rpts file to move
NOTE: These points will NOT change in the shrink wrap
NOTE: They will not be dilated or moved in min_cylinder
-v: verbose
-I #: save vertex files and (if SAVE_ENERGY defined in min_cylinder)
files of the energy of the vertices every # iterations.
These can be read in and displayed by ~jac/bin/wire
ACTIONS:
shrink_wrap takes a mesh of points and fits it to some
surface. To initialize the mesh, use planimeter to outline
the cell in a few key z slices, include at least the first and
last z slices, as well as any key features (bulges, bends, etc).
To create an attractant function, a default Gaussian psf is used.
This psf is located at /storage/big1/lml/Linux/bin/psf8.i2i.
If this is compressed, make a copy in your favorite dir, uncompress
and tell shrink_wrap where it is whith the -P option.
NOTES:
Input: this shell script expects the length of the cell to be
in the Y direction. An rpts file should be created from a copy
of the image created with transp -YZ file.i2i file_T.i2i.
(where file.i2i is your image file, and file_T.i2i is your
image file transposed in Y and Z).
Output: three intermediate files will be created (and deleted), one called output_root.i2i
which contains the coordinates of the final deformed mesh,
output.rpts which will contain an rpts file with a surface
drawn from output.i2i, and output__root_B.i2i which is the blurred input image
source code is in /storage/big1/lml/jac/was_invitro/Projects/shrink_wrap
EXAMPLE:
suppose you have a cell cell.i2i that has its length in the Y direction.
and you want to save the cell output as cell_YZ_shrink.rpts:
transp -YZ cell.i2i cell_YZ.i2i
planimeter3 -I cell_YZ.i2i -R cell_YZ.rpts
shrink_wrap -R cell_YZ.rpts cell cell_YZ_shrink >& cell.log
cell_YZ_shrink.rpts will be created (along with intermediate files cell_YZ_shrink* which get deleted).
The messages which result will be located in cell.log
dave -I cell.i2i -T 1 YZ cell_YZ.rpts cell_YZ_shrink.rpts
(due to a bug you may have to zoom out of the image a bit for the rpts data to show up on dave)
addlines cell_YZ.i2i cell_YZ_shrink.rpts - | play -P -1 R -
SEE ALSO: Corecell.pl (to apply after shrink_wrap)
Correct Orientation of the cell
______________________________ _____________________________
| . . | | . . ...... |
| . . | | . . . |
| . . | | . . |
| . . | | . . |
| . . | | . . |
| . . | | . . . |
| . . | | . . |
| . . | | . . |
| . . | | ............ |
|_____________________________| |___________________________|
length of cell in Y direction cell after transp -YZ
correct orientation for input correct orientation for drawing
file outlines with planimeter
main_memory: Command not found.
Eval: Command not found.
snakeTrack
Tracking a 2D time series using snakes.
The program takes as input an image (a 2D timeseries)
and a starting contour for the first slice of the image
in .rpts or .con format.
The program will attempt to track the moving contour
from one slice to the next.
It does this by mimimizing
F = I - E where I is the internal energy of the snake and
E is the external energy as represented by the input image
The internal energy is defined as the similarity to the original
shape of the starting contour. If a con file is specified as
input (instead of rpts file) it is recognized as such automatically
by the magic number at its top (see ~dal/synapse/projects/snakeTrack for an example).
Con files are 5-tuples: x y alpha beta lambda where alpha and
beta are shape descriptors (which can be generated automatically
by rpts2con). Lambda is a weight on the local internal energy of deformation
and 1-lambda is the weight on the local external energy.
The program will produce as output a
.rpts file, which will contain an object for each slice.
The objects can then be superimposed on the original image
using addlines.
A contour in .rpts format can be converted into the native
GSNAKE .con format using rpts2con.
For more information on snakes refer to
http://www.cs.wisc.edu/computer-vision/projects/gsnake.html
Usage :snakeTrack <image> <input rpts|con> <output rpts> [option]
[ option ]
-V :verbose - show the multiresolution pyramid.
-L# :Number of levels of resolution, default 2
-S# :Stratified search segment spacing, default 2
-N# :The size of the neighbourhood around
a snaxel to be searched, default 2
-D# :Desired spacing between snaxels.
Default 5. The program will try to keep
the spacing constant by inserting new snaxels.
It doesn't seem to do a good job deleting extra snaxels.
-E/i/e/m :External energy input type, default e
i - Use intensity as external energy value
e - Use edge magnitude and direction calculated
automatically from the input image.
m - Use edge magnitude only, calculated automatically
-slice # # :specify 0-based index of the first and
the last slice of the sequence
Note: options (L, S, N, D) must not have a space before arg.
Keywords: deformable model, active contour.
thin3Ds
This program is for thinning (i.e., finding the skeleton or medial axis) 3D
objects. It is modified from Lee, Kashyap, and Chu (a parallel algorithm).
The program attempts to thin the object, leaving only "center" voxels.
It peels off outer layers until just the "medial" axis is left.
Then the program converts the thinned image into a set of vectors in rpts format.
The motivation behind this program is for creating thin lines (vectors)
from an image data set. These lines stay thin even when zoomed way up in DAVE.
This program is similar to thin3Dp but uses a sequential implementation rather
than the parallel implementation of thin3Dp. This version has no known bugs.
Diagnostics (logfile info) are printed to stderr.
Usage: thin3Ds [options] image.i2i vectors.pts
-t #: pixels above this are foreground (default = 0)
-n name: name of vectors put into vectors.pts. The default = pixel_vectors
DAVE requires objects in each pts file to have a different name.
-r: only keep rings. all side_spurs should be removed.
-X low hi: only examine the image from low to hi in x (0 indexed)
-Y low hi: only examine the image from low to hi in y (0 indexed)
-Z low hi: only examine the image from low to hi in z (0 indexed)
-i filename: create an i2i image of the center voxels.
use this if you want to do further pruning (via "prune" command)
options for debugging:
-s rootname freq: save intermediate images to the specified file (rootname).
save them with a frequency of freq (1=save every, 2= every other, etc.
-p pass: save all the subpasses of this pass.filnames will be:
"pass#_subpass#.i2i" unless -s is also specified in which case
they will be "rootname#_subpass#.i2i"
note: if you also want the -s option, you must specify it AFTER
this option on the command line.
example:
thin3Ds -i data_center.i2i data.i2i data_thinned.pts
prune data_center.i2i data_pruned.i2i
im2pts -n data_pruned data_pruned.i2i data_pruned.pts
DAVE -n 0 -I data.i2i data_pruned.pts
note: The only program which will correctly read in the pts file produced is DAVE.
This is because many vertices in the file should not be connected to their
previous vertex, and only DAVE correctly recognizes this.
You MUST use the -n 0 option with DAVE when reading in the pts file produced.
See also: distance_transform followed by max_planes and im2pts
see also: thin (a distance based thinning algorithm)
lee94-3dskeleton.pdf (the algorithm which thin3Ds implements).
ImageJ (Fiji -> Plugins -> skeletonize 2D/3D which is an implementation of the same algorithm
http://imagejdocu.tudor.lu/doku.php?id=plugin:morphology:skeletonize3d:start
http://imagejdocu.tudor.lu/doku.php?id=plugin:analysis:analyzeskeleton:start
see also: prune
Source code in /home/lml/krypton/was_big1/was_invitro/Doxsey
/home/lml/krypton/bin/thin3Ds
track
#/home/lml/krypton/bin/track
Thresholds inimage.i2i. Connected voxels define regions.
Tracks regions over time (if -n specified). Tracks written to stdout in rpts format.
A 1-pixel border around the image is set to background.
Usage: track [options] inimage.i2i
options:
-a #: which match algorithm to use. 1: simple. 2: greedy (better)
3: min cost (best). default = 1.
-C #: what cost algorithm to use for track terminations (merges) and starts (splits).
The default is 1, constant high cost. 2 means smaller regions
incur less cost.
-p: add a penalty for a match which displaces a great match (only with -a 3)
-n #: number of sets (ie, time points in a 4D data set)
-m # #: allow paths to merge (or split) if manhattan distance between bboxes of
the regions is less than # and iod change is less than # (5.000000 and 0.200000 good values?)
-s # #: which sets to analyze (0 indexed, inclusive). must specify -n with this.
default is all sets.
-t #: an object has voxels > #. default = 0
-S: autoscale each time point to be between 0 and 255. threshold is applied after this.
this lets one compensate, eg, for bleaching over time and makes the slider value shown
by DAVE when it autoscales directly applicable to track.
-v # #: an object has between # and # voxels in it, inclusive, ie, object size. default = 1 10000
-c #: connectivity. #: 4 (2d), 8 (2d), 6, 18, 26 (3d). default = 26
-o # # outimage.i2i: write out time points # through # (0-indexed,inclusive) with voxels numbered
according to their track. note: should not need to use imsets program afterwards
on outimage.i2i. You can give numbers outside zdim (ie, -o -1 1000 would insure all
time pts save regards of numsets)
-O # # outimage.i2i: same as -o, but writes out original voxel values (voxels not in objects set to 0).
-r: print extra info (to stdout) about each region found.
-d # #: maximum delta x (or y) (in voxels), max delta z for an allowed match along a track
default=any.
-u # # # #: units for velocity measurements. x y z voxel size (um) and delta t (in sec).
-h # file: bin width (um/sec) and file name for velocity histogram (must specify -u). defaults =
0.010000 (um/sec) and vel_hist.xy
-M : also print cube outlines of the maxima in the regions (in rpts format) to stdout.
-i image.i2i: write out costs used as an image (max truncated to 32767). For diagnostic purposes.
x axis in region at time t, y axis is region at time t+1.
-V : verbose
examples:
track -t 660 -n 100 -s 0 5 wt1_r.i2i > tracks.rpts
dave -4 -I wt1_r.i2i -kptcolor -n 0 -u tracks.rpts
track -t 660 -n 100 -o -1 1000 wt1_obj.i2i -s 0 5 wt1_r.i2i > tracks.rpts
dave -4 -I wt1_obj.i2i -kptcolor -loadmap random.map -n 0 -u tracks.rpts
source code in ~lml/voxel/Corvera/track
track3d
#/home/lml/krypton/bin/track3d
Reads in mulitiple 4D images. Thresholds the images. Connected voxels define regions.
Tracks regions over time (if -n specified). Tracks written to stdout in rpts format.
A 1-pixel border around the image is set to background.
All images must be the same xdim, ydim, and zdim (after divison by tdim). tdim can vary from image file to image file.
Image files must have their tdim set correctly (see imsets program).
Usage: track [options] inimage.i2i [inimage2.i2] ... [inimageN.i2i]
options:
-a #: which match algorithm to use. 1: simple. 2: greedy (better)
3: min cost (best). default = 1.
-C #: what cost algorithm to use for track terminations (merges) and starts (splits).
The default is 1, constant high cost. 2 means smaller regions
incur less cost.
-p: add a penalty for a match which displaces a great match (only with -a 3)
-n #: number of sets (ie, time points in a 4D data set). Use if only 1 input file and it has a bad tdim.
-m # #: allow paths to merge (or split) if manhattan distance between bboxes of
the regions is less than # and iod change is less than # (5.000000 and 0.200000 good values?)
-s # #: which sets (time points) to analyze (0 indexed, inclusive, concatenated across all input files).
default is all sets. Note, currently all sets before start set are read and then
discarded, so it might be slow to skip many sets (time points).
-t #: an object has voxels > #. default = 0
-e numbkgnd hwidth delta ptsfile: rather than find regions by just thresholding, find regions by finding maxima.
A maxima still has to be above the threshold (-t) in intensity, and must be at least
"delta" greater in intensity than its neighbors (e.g, .5). The halfwidth of the
neighborhood is "hwidth" (ie, 1 means +/- 1 pixel). A pixel is also not allowed to
be a maxima if more than "numbkgnd" neighboring pixels are background pixels (ie, <= threshold).
If "ptsfile" is not "none" a pts file of the specified name will be written of the maxima.
zdim should be >> hwidth if it isn't = 1 (i.e., if it isn't a 2D time series).
-S: autoscale each time point to be between 0 and 255. threshold is applied after this.
this lets one compensate, eg, for bleaching over time and makes the slider value shown
by DAVE when it autoscales directly applicable to track.
-v # #: an object has between # and # voxels in it, inclusive, ie, object size. default = 1 10000
-c #: connectivity. #: 4 (2d), 8 (2d), 6, 18, 26 (3d). default = 26
-o # # outimage.i2i: write out time points # through # (0-indexed,inclusive) with voxels numbered
according to their track. note: should not need to use imsets program afterwards
on outimage.i2i. You can give numbers outside zdim (ie, -o -1 1000 would insure all
time points are saved regardless of numsets)
-O # # outimage.i2i: same as -o, but writes out original voxel values (voxels not in objects set to 0).
-r: print extra info (to stdout) about each region found.
-d # #: maximum delta x (or y) (in voxels), max delta z for an allowed match along a track
default=any.
-u # # # #: units for velocity measurements. x y z voxel size (um) and delta t (in sec).
-h # file: bin width (um/sec) and file name for velocity histogram (must specify -u). defaults =
0.010000 (um/sec) and vel_hist.xy
-M : also print cube outlines of the maxima in the regions (in rpts format) to stdout.
-i image.i2i: write out costs used as an image (max truncated to 32767). For diagnostic purposes.
x axis in region at time t, y axis is region at time t+1.
-V : verbose
examples:
track -t 660 -n 100 -s 0 5 wt1_r.i2i > tracks.rpts
dave -4 -I wt1_r.i2i -kptcolor -n 0 -u tracks.rpts
track -t 660 -n 100 -o -1 1000 wt1_obj.i2i -s 0 5 wt1_r.i2i > tracks.rpts
dave -4 -I wt1_obj.i2i -kptcolor -loadmap random.map -n 0 -u tracks.rpts
see also:
trackinfo.pl, modtracks.pl, ~lml/krypton/Arndt/test/run1
old source code in ~lml/voxel/Corvera/track
source code in ~lml/krypton/track
track_back2
#/home/lml/krypton/bin/track_back2
error: -readmax MUST be specified on the command line
Takes paths produced by exo4 or exo5 and findpaths_exo4.pl (or 5) and tracks back from the fusion
event as far as possible. It ignores the positions of the path found, just looks backwards for maxima
within the -diststill distance of the fusion event. A maximum being gone for at least -nomaxtime
time pts defines the start of the path (as when the maximum first disappeared going backwards in time)
Prints info about how long these paths are stationary to stdout.
This would normally be run with the same options as exo4 when it created oldpath.txt
(excluding options which aren't relevant since the fusion time is already found).
usage:
track_back2 [options] image.i2i oldpath.txt
options:
-readmax filename: read maxima from filename. This is a REQUIRED "option"
-diststill # :how far (in x or y) in pixels a maxima can move and still be considered "still", default = 2
-nomaxtime # :maximum number of consecutive time pts a maximum can be missing along the path, default = 3
-iodsize # :check iod within +/- range pixels., default = 6
-iod min max :object iod (sum of voxels > threshold within iodsize region) must be within these numbers
(inclusive) while a maximum is present. defaults: min = 1, max = 1000000
-size min max :object size (number of voxels > threshold within iodsize region) must be within these numbers
(inclusive) while a maximum is present. defaults: min = 1, max = 100000
-t # :voxels must be > # to be counted as part of object. default = 0
-r filename :rpts file which specifies a mask outline.
-paths filename : list of which paths in oldpath.txt to examine (as produced by findpaths_exo4
-stationary_time option). The default is to examine all "saved_paths" in oldpath.txt
-T startt stopt : only examine these time points (0-indexed, inclusive). may not work.
-debugpath #: number of path to debug. can only be set once.
-verbose
example:
exo4 -v -r mask.rpts -d 2400 -w 100 -jump 20 -readmax max.rpts
-iodsize 6 -maxspeed 1 -diststill 1 -fall 11 0.5 -iodrise 0.1 -minpeakval 50
-minpeakratio 2 2 3 -primedtime 10 100 -check_max 50 image.i2i >& file.out
findpaths_exo4.pl -saved_paths -stationary_time=file.stat -rptsfile=file.rpts -printfusion=5 file.out
addlines file.rpts image.i2i -|play - examine events, edit file.stat to just keep good events.
track_back2 -r mask.rpts -readmax max.rpts -iodsize 6 -diststill 1 -verbose
-paths file.stat image.i2i file.out > file2.stat
track_pts
This programs takes a list of initial pts in a 3D image and tracks
them over time. It writes out the new list. The image.i2i file should
be a 3D time series. The format for input.coords is coordinate triplets, one
triplet per line, e.g.:
x1 y1 z1
x2 y2 z2
Output is in a similar format but each point has all its time points printed
in order, along with the pixel value at each location and the average of the 3 by 3 by 3
neighborhood around the point. Points are separated
by a comment line (# at the beginning of the line)
Tracking is performed by finding the brightest point in the neighborhood of the point's
previous location in the time series.
By default, pixel locations are assumed zero-indexed (first pixel is 0,0,0).
The maximum # of pts allowed is 1000, the time series must be less than 400 in length.
Usage: track_pts [options] image.i2i input.coords output.coords
options:
-x width: change the default neighborhood halfwidth to search in x (default = 3)
-y width: same for y
-z width: same for z
-o: pixel coords in input.pts are one-indexed (like "play").
coords in output.pts should also be one-indexed.
-m: Use this when the initial coords in input.pts are not exact.
The program will move them to be the location of the brightest nearby voxel.
-c: perform hill climbing. Instead of just going to the brightest point
in the neighborhood, go to the brightest point in the neighborhood
which can be gotten to by CONTINUOUSLY going uphill. Without the -c
small intensity dips (eg, between mitochondria) could be skipped over.
-i: do not allow a tracked point to move outside the neighborhood of
it's INITIAL position (the default is the neighborhood of it's position
in the previous time frame). note: the -m option is applied
prior to this option (ie, results of -m define the "initial" position).
-b x y z: also include, as the last "point" printed out, the average background
around the point (x,y,z). The halfwidth of the region can be set with -X,Y,or Z
The coords are 0 indexed unless the -o option is also on the command line.
-X hwidth: change the default halfwidth of the background region around (x,y,z)
default = 5
-Y hwidth: same for y.
-Z hwidth: same for z.
-v: verbose. prints extra info to stderr.
-t tdim: don't read tdim from image header, just use this value.
note: entire image name (with extension) is NOT required.
note: a - in place of an image name means stdin or stdout.
note: a compressed image will be automatically be uncompressed.
see also: coords2pts.awk and coords2xmgr.awk to convert the output.coords into
either pts format (for addlines or DAVE) or xy format for xmgr.
coords2pts.awk will be high by 1 if you use the -o option in track_pts.
trackspots: performs cross-correlation before calling track_pts.
display_tracks: displays output.coords in DAVE showing intensity of tracks.
Souce code in: /storage/big1/lml/was_invitro/Drummond
vcspat
Usage:
vcspat [options] ascii_output.data mask_image.i2i image.i2i [second.i2i]
Description:
Complete spatial randomness of punctates in a volume.
Options:
-c # : do colocalization instead of complete spatial randomness
does the random trials by randomizing the points in the second image.
(use this option with two images, plus it keeps all the points in mask for
both images)
-d # : maximum distance from punctate to examine (default=10.000000)
-n # : number of regions to examine (default=100)
if # = 0, all regions in each object will be examined.
-m # : value of region in mask (default=1)
-r # : seed for random number generator (defaults to process id).
-t # #: image number and threshold. Keep points >= to image threshold (default=1 image is 1 or 2).
-T # : number of random trials. (default=100)
-z # : z weighting factor (default=1.0).
note: entire image name (with extension) is required.
note: a - in place of an image name means stdin or stdout.
Statistics
cluster_distances.pl
Takes output from channel_current2 (after 11/24/09) and pulls out statistics about
mean distance to each BK cluster. Then produces average stats from all the files
analyzed. Output to stdout.
usage:
-files:file1:file2:...:filen which files to analyze. Defaults are files named 2_*.currents in cwd.
-numBKvsProb also prints numBK vs prob data suitable for xmgrace
example:
(channel_current2 .... > file.channel_current) >& file1.currents
(channel_current2 .... > file2.channel_current) >& file2.currents
cluster_distances.pl -numBKvsProb -files=file1.currents:file2.currents > avg_distances.txt
fgrep -v clusters avg_distances.txt |xmgrace -pipe
note: mean distances to clusters are not always increasing (e.g., mean distance to 10th
cluster could be SMALLER than the mean distance to the 9th cluster). I think this
has to do with limiting my maximum distance (see comments in this file).
see also:
channel_current2
/home/lml/krypton/Zhuge/BKandRyr/analysis6/run12
/home/lml/krypton/Huang/Kth_pt.m
p. 637 of Cressie.
coloc.pl
calls coloc3way with a bunch of different thresholds. Creates an output file which can be
graphed with VennDiagram.m.
coloc.pl im1.i2i im2.i2i im3.i2i > out.coloc3way
options:
-time_vs_thresh just generate a file which shows which times correspond to which thresholds. images can be dummy names.
-t1=#:#:# threshold one should go from # to # by #. default = 800:1200:50
-t2=#:#:# threshold two should go from # to # by #. default = 800:1200:50
-t3=#:#:# threshold three should go from # to # by #. default = 2000:4000:200
-numthresh=#1:#2:#3 ignore the third number in the -t1, -t2, and -t3 options (or in the default) and instead calculate a
threshold step size so that #1 thresholds are applied to im1.i2i (dat image)
#2 thresholds are applied to im2.i2i (ctx image), #3 thresholds are applied to im3.i2i (rin image).
e.g., -numthresh=5:10:10 will apply 5 different thresholds to im1.i2i, and 10 thresholds to im2.i2i and im3.i2i.
Using this option lets me insure that each call to coloc.pl will produce exactly the same number of data points;
this may make the statistical comparison between cells easier.
-Melikian=file in addition to creating output for VennDiagram, create text file with the coloc percentages
Dr. Melikian is interested in. Graph via xmgr -nxy. See header in file for coloc info.
For this to work the three image files MUST be specified in the order 405 594 488 (i.e., rin.i2i ctx.i2i dat.i2i)
-Melikian2=file Same as -Melikian, but dat (488) is image 1, ctx (594) image 2, and rin (405) image 3
-area=file.xyr also print out Melikian data as xyr where r is proportional to the number of pixels involved
(i.e., the denominator used when calculating the percent colocalized).
prints to "file.xyr". Must also specify either -Melikian or -Melikian2
xmgrace -graphtype fixed -settype xyr file.xyr
-matlab=file:# like -area but as columns so easy to read into matlab: t p1 n1 p2 n2 p3 n3 p4 n4
# is the max n value (ie, all n values are scaled so the max over n1,n2,n3,n4 is #).
Since p1,p2,p3,p4 are percent, # is typically set to a small part of that, e.g. .05.
If # is negative then abs(#) is just used as a scale factor (so -1 would just keep the
n values as the number of voxels). Graph in matlab with line width of 2*n1:
matlab # import data=(tdim,9)
>> lowy = data(:,2) - data(:,3); hiy = data(:,2) + data(:,3); x = data(:,1);
>> area(x,hiy,'FaceColor',[1 0 0],'EdgeColor','r','LineWidth',1); hold on;
>> area(x,lowy,'FaceColor',[1 1 1],'EdgeColor','r','LineWidth',1);
Must also specify -Melikian or -Melikian2.
-debug
-verbose
example:
coloc.pl im1.i2i im2.i2i im3.i2i > coloc.data
coloc.pl -time_vs_thresh im1.i2i im2.i2i im3.i2i > coloc.times
matlab
>> VennDiagram('coloc.data','percent') # and manually look at coloc.times while doing this to convert times to thresholds.
see also:
~/krypton/packages/fiji/Fiji.app/plugins/jacob_.jar - more traditional colocalization measures
coloc_tseries2
colocalizeL
probcircle.pl
given a sphere of diameter D, what is the prob of oberving a disk of radius r when a random plane intersects it?
prints to stdout: r prob(r) 1/prob(r)
usage:
probcircle.pl [options]
options:
-D=# default = 1
-just_prob don't print the third column
examples:
probcircle.pl | xmgrace -free -pipe -nxy -
see also: p. 249 stereology_book.pdf
/home/lml/krypton/packages/fiji/stereology_lml.template
/home/lml/krypton/Corvera/Raz/stereology/plot_results.pl
/home/lml/krypton/facil/cross_sections_of_sphere
compare2dist
This program calculates whether two amplitude distributions are the same.
It was written to examine Rhonghua's STOCS data to determine whether the increase
in observed STOC frequency could be explained by an increase in STOC amplitude
(since then more events above threshold would be observed).
datafile2 should be the one with larger amplitudes (so we hypothesize it has fewer missing
events if it is just a version of datafile1 shifted higher). It will get shifted down
by a range of shifts (-s option) and compared with datafile1.
This routine also compares the temporal interarrival time distribution of each datafile with
that predicted by a poisson process and with the other datafile; the temporal calculations
ignore all missing data issues and shifting issues for now.
This program reads data in the form:
time calcium prior_baseline post_baseline amplitude
where the values are
time: time of STOC
calcium: SR calcium level
prior_baseline: amplitude just prior to STOC
post_baseline: amplitude just after STOC
amplitude: amplitude amplitude of STOC (not relative to baseline)
Data is expected to already be sorted by time (smallest to largest).
comment lines must begin with a #
Usage: compare2dist [options] datafile1 datafile2
options:
-t #: only include data with an amplitude > #. The default
is to assume a 0 threshold. If you have acquired the data
using some thresholding criterium (eg, only peaks above
10picoamps are counted) then you should provide that number
here, even if all your data is above this (ie, you've applied
the threshold in your identification phase). This is because
we need to know it when we shift amplitudes of the second
distribution to see if it could match the first distribution.
-b: subtract average baseline from amplitude prior to use (ie, prior
to any thresholding or intensity shifting of data).
-s delta max: shift datafile2 amplitudes down by a maximum of max, in
increments of delta, to see if it will then match datafile1.
-p #: our guess of the percentage of data missing from datafile2 because it was
below our threshold/detection criteria (a # from 0 to 1).
This plus the amount of datafile2 shifted below threshold determines
our estimate of the amount of data missing from the shifted datafile2 and
the unshifted datafile1. The larger this number is, the more likely
the two curves will appear similar (since differences in actual data
examined will be proportionally smaller differences, and we are assuming
all missing data matches exactly, but see -m below). Default = 0.000000
-m: assume all missing data maximally MISmatches, this would probably be
a worst case scenario, so that if the KS statistic says the distributions
could be the same even with the -m option, its probably true. But I'm
not certain of this, since it really depends how this affects the
relative spacing between the cumulative distributions. I'd have to
think about it some more.
see also:test_distr, poisson
This code is in ~lml/vision/Rhonghua/STOCS
#/home/lml/krypton/bin/compare2dist
compare2gaussian
incorrect command line, need 1 data files.
This program calculates whether a given set of data could be samples from a known mixture of Gaussians.
datafile should have one data point per line. comment lines begin with #.
Output statistics to stdout.
Usage: compare2gaussian [options] datafile
options:
-t #: only include data with an amplitude > #. The default is to include all data.
-g # amp1 mean1 sd1 ... :
The Gaussian mixture model the data should be compared against (using a KS statistic).
# is the number of Gaussians, this is followed by the amplitude, mean, and sd of
each Gaussian. The default is one Gaussian with mean = 0 and sd = 1.
You can get these values using the fitgaussians2 program.
-v: verbose. Also prints the observed (first) and theoretical (second) curves to stdout.
-T: test. runs through x = 0 to 10 and prints out cumulative prob. exits.
See Also: fitgaussians2, fit2gaussians, fit3gaussians
Source code is in ~lml/krypton/Lefkowitz
#/home/lml/krypton/bin/compare2gaussian
compare_dist2
This program calculates whether two distributions are the same using chi-square test (unless -k).
It was written to examine the distribution of sizes of Ryanodine Receptor objects
to determine if they are randomly distributed. datafile1 should be the reference
data (gotten from many simulations or from a theoretical formula), datafile2
should be the real data (max number of data values = 100000).
This program reads data which is just a list of sizes of objects seen (one per line).
Comment lines must begin with a #
Floating pt numbers are truncated to integers for bin number (max bin number = 50000).
Usage: compare_dist2 [options] datafile1 datafile2
options:
-k perform K-S test instead (I'm not certain the numbins I use to calculate this is correct).
-a also print the histograms produced
-b also print the normalized histograms (total count datafile2 scaled to the same count as in datafile1,
bins with 0 removed), this is only done if -k not specified (ie, only if performing chi-square test).
-c also print the cumulative prob distributions (only done if -k also specified)
-r # #: only analyze sizes within the specified range (inclusive)
-m #: cap the maximum number of counts for any size in datafile1 to #
-M #: cap the maximum number of counts for any size in datafile2 to #
-z: for chi-square keep zero entry bins (chi square only correct if this option is NOT used)
so why is this option here? if you just really want to use this program to do
histogram normalization (-b), and don't care about the stat. xmgrace running avg
will produce strange results if you don't use -z, ie, if data not equally spaced.
-v verbose
-V very verbose
example:
create a simulated distribution and compare it with real restored data (real_ryrr.i2i, run5 does this 100 times):
cluster -wsize 10 43 25 -cylinder 0.001 3 -psize 0.08 0.08 0.25 -mindist 0.030 -numryr 500 -image ryr.i2i
blur3d -S 40000 -P ryr.i2i psf.i2i ryr_b.i2i
noise -c RCA ryr_b.i2i ryr_bn.i2i
setenv myloc /home/lml/krypton/Zhuge/Ryr_clusters
setenv eprloc /storage/big1/epr/queues
echo " /storage/big1/epr/run_epr -V -log -NSC 4 -sm 1e-05 -it 5000 -sc 0.025 -co 0.001
-psf ${myloc}/psf.i2i ${myloc}/ryr_bn.i2i ${myloc}/ryr_bnr.i2i " > /storage/big1/epr/queues/standard/lml.0
while (1==1) if ( ( ! -e ${eprloc}/standard/lml.0 ) && ( ! -e ${eprloc}/running/lml.0 ) ) then break endif sleep 60s end
countobjs -k -s 5 300 -t 4500 4500 sim_ryr.objs ryr_bnr.i2i
cull -s -c -2 < sim_ryr.objs > sim_ryr.stats
countobjs -k -s 5 300 -t 4500 4500 real_ryr.objs real_ryrr.i2i
cull -s -c -2 < real_ryr.objs > real_ryr.stats
compare_dist2 -b sim_ryr.stats real_ryr.stats >& chi_ryr
compare_dist2 -k -b sim_ryr.stats real_ryr.stats >& ks_ryr
tail chi_ryr ks_ryr
xmgrace chi_ryr; xmgrace ks_ryr
see also: cluster, run5
This code is in /home/lml/krypton/Zhuge/Ryr_clusters/
#/home/lml/krypton/bin/compare_dist2
correlationL
/home/lml/krypton/bin/correlationL
This program plots intensities from image1 vs intensities from
image2. image1 on the x axis, image 2 on the y axis. Intensities
in both images are rescaled 0 to the size of the output image
(default = 512). The intensity at each pixel in the output
image is the number of (image1,image2) voxels with that particular
intensity combination.
The intent is to be able to look at the output image and get a sense
for whether there is a correlation between intensities
in the two images. If so, perhaps a least squares line
fit should be done to see how well one images correlates with the
other, or -s option can be specified.
The minimum, maximum, and scale factors are printed to stdout.
Version 1.1
Usage: correlation [options] im1.i2i im2.i2i out.i2i
options:
-d xdim ydim: output dimensions of new image
(default = 512,512).
-S im1scale im2scale: scale intensities with these scale factors. newval = (intens-min_intens)*scale
-B b1 b2: force min_intens (black level) to specified value. newval1 = (intens-b1)*scale1
-i val1 val2: Ignore any voxels in image1 <= val1 and
any voxels in image2 <= val2.
-h val1 val2: Ignore any voxels in image1 >= val1 and
any voxels in image2 >= val2.
-r: instead of intensities, use the rank
order of the intensities.
-a: automatically calculate the size of the
output image. attempts to make the scale
factor = 1.
-b: keep the xdim and ydim scale factors the same.
-s: calculate Spearman rank-order correlation
coefficients. print results to stdout.
-z zslice: only examine the specified z slice (the first
slice is considered slice 0).
note: entire image name is NOT required.
note: a - in place of an image name means stdin or stdout.
examples:
# to apply this to a set of 3D images, using the same scales and size images for easy comparison
correlationL -S s1 s2 -d 512 512 -B 0 0 image1.i2i corr1.i2i (and same for other images)
create_imagesL
This program takes output from many runs of "overlap" and produces two images. One image is a percent colocalization image and the other is a probability image. The overlap program MUST be run via the "many_overlaps" command. The probability calculations are the same as those performed by "probcolocal", but only -n needs to be specified since the other values are automatically extracted from the file produced by "many_overlaps". The percent image is the percent of voxels above threshold in image1 (as originally specified to "many_overlaps") which overlap with voxels above threshold in image2. A 100 in the output image = 100 percent. The probability of observing as much (or more) overlap as we did due to random chance is the prob.i2i image. Probabilities are mulitplied by 100. Position along the y-axis (vertical axis) of the image, corresponds to thresholds for image2. The x-axis represents image1 thresholds. The lowest threshold is the bottom left corner. See the first line in your datafile for the range of thresholds this represents. Overlap produces statistics for a range of allowed overlaps. Create_images, by default, chooses an overlap within 1 voxel. If you want to use a different overlap distance use the -o option. See also: overlap, probcolocal, many_overlaps Usage: create_images [options] datafile percent.i2i prob.i2i options: -o distance: voxels within this distance count as overlapped. -n number: number of voxels (1000000 default), see probcolocal. Examples: many_overlaps -t 100 1000 100 -T 100 1000 100 alpha_T.i2i beta_T.i2i tmp_output create_images -n 363264 tmp_output percent.i2i prob.i2i
makeGmixtures
This program creates a set of data which is sampled from a known mixture of Gaussians. Output statistics to stdout. This can be used to test the compare2gaussian program. Usage: makeGmixtures [options] options: -n #: number of data points to calculate, default = 100 -g # amp1 mean1 sd1 ... : The Gaussian mixture model the data drawn from. # is the number of Gaussians, this is followed by the amplitude, mean, and sd of each Gaussian. The default is one Gaussian with mean = 0 and sd = 1. Really only the relative amplitudes matter, not their absolute value since they determine the relative probability of drawing from each distribution. -v: verbose. prints extra info to stdout, each point is labeled with which Gaussian it is from. Use,e.g.,: fgrep G1 output|cull -2 > tmp.dat to then graph it. See Also: compare2gaussian, fitgaussians2, fit2gaussians, fit3gaussians Source code is in ~lml/krypton/Lefkowitz #/home/lml/krypton/bin/makeGmixtures -h
mpoisson
Generate the inter-arrival distribution seen from 10 independent poisson processes
(eg, spark sites), to check that it is equivalent to 1 process whose lambda is the
sum of the independent processes
Usage: mpoisson
options:
-t #: total time (in seconds) to count number of arrivals, default = 1000.000000
-l #: lambda of each poisson process (mean number of events per second), default = 10.000000
-p #: number of processed, default = 10
-d dur sep: drop intervals of specified duration, sep seconds between the start of one dropped
interval and the next. This is to test if the resulting process is still Poisson.
It mimics how Rhongua acquired the data (a series of disconnected 2 second intervals).
-i dt: print histogram of inter-arrival times instead. bin width is dt (in seconds)
probcolocalL
This program calculates the probability of an event occurring
k or more times in n trials if the probability of an event is
p and events are independent (see p.182 of Numerical Recipies)
It is intended as one way to determine if an observed level of
volume colocalization is due to random chance or not. To use it
in this manner, first find the fraction of voxels in each image
which are used to calculate colocalization (e.g., those above
threshold). Call these fractions f1 and f2. Then, if these
voxels were randomly placed, the probability that identical voxels
are turned on in both images is f1*f2. An "event" is just such
a coincidence, so p = f1*f2. n = number of voxels in one image.
k= number of voxels which are observed to colocalize.
This program then returns the probability that this amount of
colocalization or more would have occurred merely due to chance.
Note: if only an annulus (i.e., a subset of the total image volume)
is really being analyzed, then f1 and f2 and n should probably be
derived based on the volume subset, not the entire image.
Usage: probcolocal [options]
Output (to standard out): the probability of this or greater occuring randomly.
options:
-n number: number of trials (100 default)
-k occurrences: number of occurrences (50 default).
-p probability: prob of an event (0.500000 default).
-h: print this help message.
roc
/home/lml/krypton/bin/roc
This program reads two images. It attempts to use voxel values in
image1 as a predictor of voxel values in image2; ie, as a type of
measure of colocalization. Both images are first rescaled to be
between 0 and max (100).Then image2 is thresholded. Image1
is then set,sequentially, to all integer thresholds between 0 and
and max (unless -r is used to restrict the range). If a voxel
in image1 is above threshold then we guess
that the same voxel is above threshold in image2.
It also does the reverse, ie, use image2 to predict image1.
The default cost function is: 1- 255*prob(detection)*(1-prob(false alarm)).
OUTPUT:
TO STDOUT:
The best thresholds, lowest cost and probability of detection,
prob of failure, prob of miss and prob of saying nothing when
there is nothing.
TO A FILE:
6 images get created, they are the costs and probability of
detection and false alarm (3 separate images) when im1 is ussed
to predict im2, and another 3 for when im2 is used to predict im1.
By default these will be named:
im1predictingim2_c.i2i, im1predictingim2_pd.i2i, im1predictingim2_pf.i2i
im2predictingim1_c.i2i, im2predictingim1_pd.i2i, im2predictingim1_pf.i2i
The -p option changes the prefix (eg,the "im1predictingim2").
The images all have thresh1 on x axis, thresh2 on y axis.
The cost images are always scaled 0-255.
The pd and pf images are scaled 0-100.
Additional output files can be created with the -c option.
SEE ALSO: playims . This program quickly puts up all 6 images
created by roc. It can take arguments.
Usage: roc [options] image1 image2
options:
-m max: max of intensity range to rescale data to
the default is 0 to 100.
-b val1 val2: ignore all background voxels. A voxel will be
ignored in both images if the voxel in image1
is <= val1 and the voxel in image2<= val2.
-r low hi: range of intensity threshold values to check
in image1 (in original image intensity units),
default is entire intensity range.
-R low hi: range of intensity threshold values to check
in image2 (in original image intensity units),
default is entire intensity range.
-t thresh: only examine this threshold value for image1.
-T thresh: only examine this threshold value for image2.
I am not sure -t and -T work completely correctly.
-c file1 file2:
Creates 2 roc curve data files for graph and plot.
The probability of false alarm (x coord) and
detection (y coord) and cost (intensity) for all
thresholds in image1 for a threshold in image2
is written file1. A text string
indicating the image2 threshold level is written out.
This is then repeated for other image2 threshold levels.
file2 is the same, but for image2 predicting im1.
Use: graph -f 1 -b <file1 |plot to display these roc curves.
-C d f m n: d is the "cost" of a correct detection, ie
a voxel in image1 is above threshold the same time
that the equivalent voxel is above thresh2 in
image2.
f is the cost of a false alarm (voxel in
image1 > thresh1 but voxel in image2 is < thresh2).
m is cost of a miss (im 1 <t1, im 2 > t2) and
n is cost of nothing present ( both im 1 and im 2
less than threshold, ie correctly predicting that
nothing is there).
Given these costs the program will print to stderr
the thresh1 and thresh2 values which minimize the
total cost. It will also print the total cost.
Total cost = d*pd + m*pm + f*pf + n*pn
where pd = probability of detection, etc.
Typically, d = n = 0 and f = m = 1.
-p pre1 pre2: prefixes for the names of the saved images
which show the sensitivity analysis. 6 images
get created (regardless of whether the p option
is specified), they are the costs and prob. of
detection and false alarm (3 separate images)
when im1 is used to predict im2, and another 3
for when im2 is used to predict im1.
By default these will be named:
im1predictingim2c.i2i, im1predictingim2pd.i2i,
im1predictingim2pf.i2i, im2predictingim1c.i2i, etc.
This option changes the prefix (ie, what comes
before the "c","pd",and "pf") for the
images saved. pre1 will be for im1 predicting im2,
and pre2 will be vis versa.
-s file: save roc data as an array of floats to "file".
Once this file is saved, any particular values
of costs can be examined very
quickly. In other words, this is the data that
the roc program is really taking a long time to
calculate. So if you want to examine the same
pair of images under different cost
scenarios, save the data file during the 1st
scenario, and run the others using the saved data.
NOT IMPLEMENTED YET.
-f file: read in a saved array (see -s above). When this
option is specified the args image1 and image2 are
not specified on the command line.
NOT IMPLEMENTED YET.
note: entire image name is NOT required.
note: a - in place of an image name means stdin.
Typical examples:
1). roc im1.i2i im2.i2i > roc.info
playims dummy
2). roc -p 1to2 2to1 -b 0 0 im1.i2i im2.i2i > roc.info
3). roc -C 0 1 1 0 -b 0 0 im1.i2i im2.i2i > roc.info
4). roc -r 20 80 -R 50 200 im1.i2i im2.i2i > roc.info
5). roc -b 5 5 -c roc1to2.dat roc2to1.dat im1.i2i im2.i2i > roc.info
graph -f 1 -b <roc1to2.dat |plot
graph -f 1 -b <roc2to1.dat |plot
playims dummy
sparkstats5
#/home/lml/krypton/bin/sparkstats5 sparkstats5 is similar to sparkstats3 (only examines the first 10 seconds of data from any cell and ignores cells which don't have 10 seconds of data). It eliminates from any 2 second interval multiple events at the same site, since we guess that is due to a second "bursting" process which we want to ignore. All events in the same cell, at the same x,y are considered events from the same spark Usage: sparkstats5 [options] datafile options: -N #: spark events considered the same site if <= # pixels in x and y from first event and also in the same cell and 2 second interval (note: histogram produced does not compensate for the longer time some cells were under observation). -O #: like -N. in addition, sparks with 0 events in a 2 second interval are added for each spark that exists in an interval but does not exhibit spark events in some other interval in the same cell. -i: spark events condidered the same "site" if in same cell and same 2 second interval this is the default. -I: an extra spark id field does NOT precede the spark data (ie, test.data, not bestdata.data). -J: datafile is just one cell; data format: x y amp; just calculate spark sites (use specify -N) [source code in /usr/people/lml/vision/Rhonghua/Sparks/sparkstats5.c]
Analysis of Points
components.pl
Calculates the number of connected components resolved over a range of point densities. components.pl [options] options: -numpts=low:hi:delta -repeat=# number of times to repeat at each point density -res=# in microns -outfile=name name of output file. otherwise to _tmp. always need write permission in current directory.
convex_hull.pl
This program takes an object outputfile (unless -rptsin or -tripletsin) produced by countobjs (or maskobjs) and finds the minimal
spanning tree connecting the center of mass of the objects, and the convex hull. Writes out connections as an rpts file to stdout.
It also loops through 20 thresholds for the pts, and prints to stderr some stats about the minimal spanning tree and
convex hull of the points above each of these thresholds.
Options can be used to only selected a subset of the objects and to constrain the maximum distance (-maxdist).
It was written with the hope that triplets (maxima in an image which has a cell in the center) will show a pattern in their
convex hull area, so that when the threshold rises high enough, only maxima inside the cell will still be present and hence
the convex hull will be stable and of much smaller area.
This program requires perl modules Graph::Kruskal and Math::Geometry::Planar, use cpanp to install these:
yum install perl-CPANPLUS-0.91.3-202.fc16.noarch - for cpanp
cpanp -i Graph::Kruskal
cpanp i Math::Geometry::Planar
usage:
convex_hull.pl -tripletsin file1.triplets > file1_mst.rpts
input options:
-dx=# spacing in the x direction, default = 1
-dy=# spacing in the y direction, default = 1
-dz=# spacing in the z direction, default = 1
-rptsin instead of file1.objs, read file1.rpts where file1.rpts was produced by max2d.
-size1 and -iod1 are ignored when this option is specified.
-tripletsin instead of file1.objs read in file1.xyi, triplets of x y and intensity.
analysis options:
-size1=min:max min and max object size allowed for file1.objs in pixels (inclusive)
-iod1=min:max min and max object iod allowed for file1.objs (inclusive), or intensity if -tripletsin was specified.
-maxdist=#: : don't allow connections between pts further apart than this distance (after applying dx, dy, dz spacing factors).
warning: if the distance is too small, then all the pts will not be connected to begin with;
this may cause the minimal spanning tree algorithm to fail.
default maxdist is 3000
output options:
-addlines : output rpts format which addlines is more likely to be happy with (may not work in dave?)
-hist : calculate histogram of distances (hist[int(dist)]++, where int just truncates to integer). obsolete.
-sizehist : calculate histogram of object sizes (in voxels) in file1.out. (hist[int(size)]++, where int just
truncates to integer). If you don't care about distances between two files (ie, only want -sizehist)
put the same file name for both files on the command line. obsolete.
-rpts=file.rpts : put bounding boxes around objects found and lines between closest objects (similar to objs2bb.pl)
display this in dave eg: dave -n 0 -u -I file.i2i file.rpts (use the -n 0 and -u options)
note: may not show up in dave until you click on "scale" to force a rescale (or draw?).
-jack=max.rpts:# : create an rpts file with the maxima in each object displayed as a small jack.
# controls the size of the jack. 1=1 pixel in size, 2 = 2 pixels, etc.
-twod add this (in addition to -jack) if you want 2D jacks rather than 3D jacks.
-strlim=# : limit length of names of objects in rpts files to # characters. dave sometimes crashes if the
names are too long.
-tyz : transpose coords in yz before writing them to the rpts (must specify -rpts option)
-color=# : for the w value (color) use the specified value (use -P in play to see this as a color). default = none.
if you have no color (no w value), addlines can put one in (-P option in addlines).
diagnostics:
-silent : suppress warning messages.
-verbose
-debug
example:
convex_hull2.pl -tripletsin -jack=jack.rpts:5 -addlines -twod -maxdist=100 file1.triplets > file1_mst.rpts
addlines2 -P -1 image.i2i file1_mst.rpts - | addlines2 -P -2 - jack.rpts - | reduceima -bin 3 3 -L - -| play -P -1 R -P -1 G -
see also:
convex_hull, objs2mst.pl
objs2bb.pl, objs2dist.pl, objs2dist121.pl, closest_object, closest_voxel, countobjs, maskobjs, objs_via_ascent
resolve_pts
objs2alldists.pl
Like obs2disp.pl but calculates the distance to ALL the objects (within -maxdist), not just the closest. See ~/krypton/Zhuge/grants/AsthmaRO1/README for 5/27/11 and /home/lml/krypton/Zhuge/BKandRyr/analysis6/run18 (which did NOT use this). This program takes 2 object outputfiles produced by countobjs (or maskobjs) and finds the shortest distance from the center of mass of objects in file1.out to the center of mass of objects in file2.out. If both file arguments are the same (e.g., objs2dist.pl file1.out file1.out >file.dist), you can use this routine to find the distances between objects within an image (an object match with the same id is automatically ignored in this case so objects don't match to themselves). See also the option -skipsame. (modified from /storage/big1/lml/was_voxel/Corvera/track/objs2bb.pl) usage: objs2alldists.pl file1.out file2.out >file.dist options: -dx=# : spacing in the x direction ,default = 1 -dy=# : spacing in the y direction ,default = 1 -dz=# : spacing in the z direction ,default = 1 -size1=min:max : min and max object size allowed for file 1 in pixels (inclusive) -size2=min:max : min and max object size allowed for file 2 in pixels (inclusive) -iod1=min:max : min and max object iod allowed for file 1 (inclusive) -iod2=min:max : min and max object iod allowed for file 2 (inclusive) -maxdist=#: : don't allow matches further away than this distance (after applying dx, dy, dz spacing factors). default maxdist is ~ 3000. -hist : calculate histogram of distances (hist[int(dist)]++, where int just truncates to integer). -sizehist : calculate histogram of object sizes (in voxels) in file1.out. (hist[int(size)]++, where int just truncates to integer). If you don't care about distances between two files (ie, only want -sizehist) put the same file name for both files on the command line. -rpts=file.rpts : put bounding boxes around objects found and lines between closest objects (similar to objs2bb.pl) display this in dave eg: dave -n 0 -u -I file.i2i file.rpts (use the -n 0 and -u options) note: may not show up in dave until you click on "scale" to force a rescale (or draw?). -jack=max.rpts:# : create an rpts file with the maxima in each object displayed as a small jack. # controls the size of the jack. 1=1 pixel in size, 2 = 2 pixels, etc. -strlim=# : limit length of names of objects in rpts files to # characters. dave sometimes crashes if the names are too long. -tyz : transpose coords in yz before writing them to the rpts (must specify -rpts option) -skipsame : don't match an object in file1.out to an object with the same id in file2.out This is set automatically if file1.out = file2.out. -silent : don't print the distances (sets -hist) -simpleprint : when printing distances, just print distance, one per line, nothing else. to be read in by ~/Zhuge/grants/AsthmaRO1/distance_distribution.m. note: that program assumes distances are in pixels and pixels are 80 nm, unless you specify pixelsize=somethingelse. -simpleprint2 : like simpleprint, but the first column is id of the Ryr to which this BK is closest. -onelineprint : like objs2alldists.pl but prints the distance to ALL the objects (within -maxdist), not just the closest on one line, so we can keep track of which BK are near which Ryr. see comdists3.pl (which uses this option). -verbose example: countobjs -k -t 2000 2000 wave1.objs wave1.i2i countobjs -k -t 2000 2000 wave2.objs wave2.i2i objs2dist.pl wave1.objs wave2.objs > wave1and2.dist see also: objs2dist121.pl, closest_object, closest_voxel, countobjs, maskobjs, objs_via_ascent, pt_distances
objs2dist2.pl
objs2dist2.pl: like objs2dist.pl but when -rpts specified prints out lines to ALL objects within -maxdist (which now defaults to 20), rather than just the closest. So we can see all the BK clusters which are close enough to a Ryr to be activated. see /home/lml/krypton/Zhuge/paper2010/README for 5/20/10. This program takes 2 object outputfiles produced by countobjs (or maskobjs) and finds the shortest distance from the center of mass of objects in file1.out to the center of mass of objects in file2.out. If both file arguments are the same (e.g., objs2dist.pl file1.out file1.out >file.dist), you can use this routine to find the distances between objects within an image (an object match with the same id is automatically ignored in this case so objects don't match to themselves). See also the option -skipsame. (modified from /storage/big1/lml/was_voxel/Corvera/track/objs2bb.pl) usage: objs2dist2.pl file1.out file2.out >file.dist options: -dx=# : spacing in the x direction ,default = 1 -dy=# : spacing in the y direction ,default = 1 -dz=# : spacing in the z direction ,default = 1 -size1=min:max : min and max object size allowed for file 1 in pixels (inclusive) -size2=min:max : min and max object size allowed for file 2 in pixels (inclusive) -iod1=min:max : min and max object iod allowed for file 1 (inclusive) -iod2=min:max : min and max object iod allowed for file 2 (inclusive) -maxdist=#: : don't allow matches further away than this distance (after applying dx, dy, dz spacing factors). default maxdist is 20. -hist : calculate histogram of distances (hist[int(dist)]++, where int just truncates to integer). -sizehist : calculate histogram of object sizes (in voxels) in file1.out. (hist[int(size)]++, where int just truncates to integer). If you don't care about distances between two files (ie, only want -sizehist) put the same file name for both files on the command line. -rpts=file.rpts : put bounding boxes around objects found and lines from objects in file1.out to all objects in file2.out that are <= maxdist away (these lines will become an object named "near_to_file1.out"). display this in dave eg: dave -n 0 -u -I file.i2i file.rpts (use the -n 0 and -u options) note: may not show up in dave until you click on "scale" to force a rescale (or draw?). -jack=max.rpts:# : create an rpts file with the maxima in each object displayed as a small jack. # controls the size of the jack. 1=1 pixel in size, 2 = 2 pixels, etc. -strlim=# : limit length of names of objects in rpts files to # characters. dave sometimes crashes if the names are too long. -tyz : transpose coords in yz before writing them to the rpts (must specify -rpts option) -skipsame : don't match an object in file1.out to an object with the same id in file2.out This is set automatically if file1.out = file2.out. -silent : don't print the shortest distances (sets -hist) -verbose example: countobjs -k -t 2000 2000 wave1.objs wave1.i2i countobjs -k -t 2000 2000 wave2.objs wave2.i2i objs2dist2.pl wave1.objs wave2.objs > wave1and2.dist see also: objs2dist121.pl, objs2dist.pl, closest_object, closest_voxel, countobjs, maskobjs, objs_via_ascent, pt_distances
objs2mst.pl
This program takes an object outputfiles (unless -rptsin) produced by countobjs (or maskobjs) and finds the minimal spanning tree connecting the center of mass of the objects. Writes out connections as an rpts file to stdout. Options can be used to only selected a subset of the objects and to constrain max distance (so multiple trees can be produced). usage: objs2mst.pl file1.objs > file1_mst.rpts options: -dx=# : spacing in the x direction, default = 1 -dy=# : spacing in the y direction, default = 1 -dz=# : spacing in the z direction, default = 1 -size1=min:max : min and max object size allowed for file1.objs in pixels (inclusive) -iod1=min:max : min and max object iod allowed for file1.objs (inclusive) -rptsin : instead of file1.objs, read file1.rpts where file1.rpts was produced by max2d. -size1 and -iod1 are ignored when this option is specified. -addlines : output rpts format which addlines is more likely to be happy with (may not work in dave?) -maxdist=#: : don't allow matches further away than this distance (after applying dx, dy, dz spacing factors). default maxdist is ~ 3000. -hist : calculate histogram of distances (hist[int(dist)]++, where int just truncates to integer). -sizehist : calculate histogram of object sizes (in voxels) in file1.out. (hist[int(size)]++, where int just truncates to integer). If you don't care about distances between two files (ie, only want -sizehist) put the same file name for both files on the command line. -rpts=file.rpts : put bounding boxes around objects found and lines between closest objects (similar to objs2bb.pl) display this in dave eg: dave -n 0 -u -I file.i2i file.rpts (use the -n 0 and -u options) note: may not show up in dave until you click on "scale" to force a rescale (or draw?). -jack=max.rpts:# : create an rpts file with the maxima in each object displayed as a small jack. # controls the size of the jack. 1=1 pixel in size, 2 = 2 pixels, etc. -strlim=# : limit length of names of objects in rpts files to # characters. dave sometimes crashes if the names are too long. -tyz : transpose coords in yz before writing them to the rpts (must specify -rpts option) -color=# : for the w value (color) use the specified value (use -P in play to see this as a color). default = none. if you have no color (no w value), addlines can put one in (-P option in addlines). -silent : don't print the shortest distances (sets -hist) -verbose example: countobjs -k -t 2000 2000 wave1.objs wave1.i2i objs2mst.pl wave1.objs > wave1mst.rpts dave -I wave1.i2i -n 0 -u wave1mst.rpts see also: objs2bb.pl, objs2dist.pl, objs2dist121.pl, closest_object, closest_voxel, countobjs, maskobjs, objs_via_ascent resolve_pts
objs_within_dist.pl
This program takes 2 object outputfiles produced by countobjs (or maskobjs) and finds the number of objects in the second file which are within a given distance of each object in the first file. A histogram is printed out. Distances are from the center of mass of objects in file1.out to the center of mass of objects in file2.out. (modified from objs2dist.pl) usage: objs_within_dist.pl file1.out file2.out >file.dist options: -dx=# : spacing in the x direction ,default = 1 -dy=# : spacing in the y direction ,default = 1 -dz=# : spacing in the z direction ,default = 1 -size1=min:max : min and max object size allowed for file 1 in pixels (inclusive) -size2=min:max : min and max object size allowed for file 2 in pixels (inclusive) -iod1=min:max : min and max object iod allowed for file 1 (inclusive) -iod2=min:max : min and max object iod allowed for file 2 (inclusive) -maxdist=#: : don't allow matches further away than this distance (after applying dx, dy, dz spacing factors). default maxdist is ~ 3000. -rpts : read in file in rpts format -verbose : extra info written to stderr -vverbose : print other info. can be combined with -verbose. example: countobjs -k -t 2000 2000 wave1.objs wave1.i2i countobjs -k -t 2000 2000 wave2.objs wave2.i2i objs_within_dist.pl wave1.objs wave2.objs > wave1and2.dist see also: objs2dist121.pl, closest_object, closest_voxel, countobjs, maskobjs, objs_via_ascent, pt_distances
connected_random_pts
Generates random pts in a planar region, calculates how many connected components are created if a pt is connected to all other pts within a given radius. Usage: connected_random_pts [options] Options: -x lowx hix: pts have xcoords within this range, default = (0.0,100.0) -y lowy hiy: pts have ycoords within this range, default = (0.0,100.0) -r #: radius (in pixels), default = 5.0 -s #: long int starting seed for random number generator. default = process id. -n #: number of pts to create, default = 1000 -h: help -v: verbose See also: krypton/Huang/README, components.pl, Kth_pt.m, num_observed.m, mean_minimum_distance.m,objs2mst.pl Source code in ~krypton/lml/facil
pt_distances
#/home/lml/krypton/bin/pt_distances
Takes output from 2 runs of max2d_simple -p file.rpts and calculates minimum distance between pts.
See comments printed to stdout when the program runs to explain the fields in the output data.
Usage:
pt_distances [options] file1.rpts file2.rpts
Options:
-h: print histograms of distances. entry for pt at dist d: histogram[int((d+tiny)/binsize)]++
-t #: set tiny to #, default = 0.000010 . Points at exactly 1 pixel seem to be put into the .9 bin
perhaps due to slight roundoff error. By adding tiny, they go into the next bin.
-b #: set binsize to # pixels, default = 0.100000
-m #: set maximum distance histogram will go out to (distances > this get put in last bin)
default = 50.000000 pixels
-a #: the area (in pixels) of the region used to generate file1.rpts and file2.rpts
this is used to compare observed rates with theoretical rates if randomly distributed.
NOTE: the theoretical calculations for image1 vs. image2 (and vice versa) are probably WRONG.
default = 10000.000000
-M #: the minimum distance allowed between maxima, for theoretical calculations. This might be
1 (pixel) since maxima normally can't be next to each other. Or it might be the
resolution of the microscope (since objects closer than that will appear as one maxima).
by default M from 0.0 to 3.0 by 1.0 is calculated. if you want an additional value use this option.
-v: verbose
See also: max2d_simple read_hist.pl
Example:
max2d_simple -t 10 -d .5 clathrin.i2i > clathrin.pts
max2d_simple -t 10 -d .5 glut.i2i > glut.pts
pt_distances -h -a 26443 -M 1.5 clathrin.pts glut.pts > both.dat
fgrep "image1 only" both.dat | fgrep mean_min | cull -n -15 -21 -24 -30 > clathrin_mindist.xy
xmgrace -nxy clathrin_mindist.xy &
read_hist.pl -im1 -norm=1 -nummax=26443 -avg=5 -nozero < clathrin_mindist.dat > clathrin_histo.xy
xmgrace clathrin_histo.xy &
Source code in ~krypton/lml/facil
resolve_pts
Takes minimimal spanning tree rpts file (just an rpts file with each pair of pts which are to be compared specified as a line segment, i.e., two pts and a "break") as produced by objs2mst.pl and finds which connected pts are resolved and which are not. Resolved pts are those which have at least the specified intensity dip along the minimum cost (roughly: brightest) path connecting those pts. note: if there is no path between the two pts such that all voxels are greater than the threshold, the pts are also considered to be resolved. If a pair of pts is resolved, both are written to out.rpts (but not more than once). If a pair of pts is not resolved then the brighter of the two (voxel intensity) is written to out.rpts. The intensity dip is the max amount the intensity goes below the dimmer of the two endpts (anywhere along the minimum cost path). Each pair of lines (i.e., pair of coordinates) in mst.rpts defines ends of a minimum cost path along which an intensity dip will be searched for. Usage: resolve_pts [options] image.i2i mst.rpts out.rpts options: -t #: threshold, default = 0.000000 -f : normally a pt is discarded if it does not resolve against at least one other pt it is connected to in the input rpts file. Thus, AXXXXBXXXXC if A and B resolve, but B doesn't resolve against C, and B is smaller than C B will normally be discarded (even though it resolves against A, it fails against C). If you specify -f then maxima which fail will be kept as long as they resolve against some other pt. -d #: intensity dip required to be resolved. default = 10.000000 -p #: the percentage dip required relative to the dimmer endpt (e.g. 26 is 26 percent dip) -l: instead of dip being the maximum amount below the dimmer of the two pts, dip is the max amount below a linearly interpolated intensity between the two endpoints. -i: truncate coords written to out.rpts to integers (addlines prefers this I think). -v: verbose. -V: very verbose. Examples: countobs image.i2i > image.objs obs2mst.pl -maxdist=10 image.objs > image_mst.rpts resolve_pts -d 100 -i image.i2i image_mst.rpts image_maxima.rpts See also: BestPath, objs2mst.pl, krypton/Huang/TIRF4/README Source code in krypton/facil
Image Manipulation
align_quadrants.pl
This program displays the four quadrant images of a well. The user then saves 3 pairs of points (s when middle mouse is pressed). Each pair of points should be points that match in two neighboring quadrants. When done, each quadrant must have one (or two) points specified. Points must be in pair order (see recalc_quadrant2.pl). When done, the use must write them out via z (when middle mouse is also pressed, no confimation is given in play). The quadrant images are then aligned based on these points and newimage.i2i is created. It will also create a file with the output from play, for future reference. It will be named, eg, G02_combined_play.pts. It can be passed manually to recalc_quadrant2.pl to redo the alignment if necessary. usage: align_quadrants.pl [options] well newimage.i2i examples: align_quadrants.pl [options] G02 G02_combined.i2i options: -no_pts_output don't produce the pts file -noblend by default the quadrant images are blended together where they overlap. use this option to turn that off. -print_overlap also print the amount of overlap to stderr -debug -debug_all see also: combine_images recalc_quadrant2.pl /home/lml/krypton/Corvera/Raz/README 5/16/14 http://wiki.imagej.net/Grid/Collection_Stitching_Plugin
combine_images.pl
This program runs combine_images on all the images in the current directory. All images which have names
like L##_morestuff.i2i (where L is any letter and # is any digit) will be combined;
all the A02 will be combined (to produce A02_comb.i2i), all the A03, etc.
The resulting image for the combination of the A02 images will be A02_tag.i2i (see -newtag option, e.g. A02_comb.i2i).
Any images already existing in the current directory with a name like L##_stuff_oldtag.i2i will be ignored.
usage:
combine_images.pl [options]
options:
-newtag=something The output filename for L##*.i2i will be L##_newtag.i2i. Default = comb
-oldtag=something Any files of the form L##*_oldtag.i2i will be ignored. Default = comb
-pixsize=# pass this as -p # to combine_images program (pixel size).
Specify -pixsize=2.41 (um/pixel) for the 2.5X objective.
-reduceima=#:#:tag After creating output image, apply reduceima to it
with -bin # # and the specified output tag on the name.
So, e.g., A02_comb.i2i will become A02_tag.i2i
-flipx flip images in X prior to combining them
-flipy flip images in Y prior to combining them (can be used at the same time as -flipx)
-x=#:#:#:# manually specified picture offsets. see combine images
-y=#:#:#:# manually specified picture offsets. see combine images
-o=# number of pixels the images overlap (ignores info in image headers). see combine_images
-f do MOT pad images at border (as of 2/3/14). see combine_images.
-remove Remove the full-size combined image (e.g., A02_comb.i2i) when done (use with -reduceima).
-dummy Just echo what would be done, but don't do anything
-verbose Echo as things are done.
-debug
note:
combine_images (in /storage/big1/lml/Linux/bin) and reduceima (in /storage/big1/kef/Linuxbin) must be in your
path for this perl script to work. You can check this by just typing: which combine_images and which reduceima;
if they are not found you need to add their directory to your PATH environment variable (echo $PATH will show you
what it is currently). How this is done varies depending upon which shell you are running. Under tcsh it would
be setenv PATH $PATH:/storage/big1/kef/Linuxbin:/storage/big1/lml/Linux/bin .
example:
cd /storage/big3/kdb/Zeiss/20130326/GEDMAT_002_PRO_SQ2_DAY14
# Combine all images in each well. The combined image will be L##_combined (-newtag=combined).
# Then reduce the combined size back down to the size of one original image;
# call the new image L##_combined_reduc.i2i (-reduceima=2:2:reduc).
# Finally, remove the big (unreduced) combined image (-remove).
# First do a dummy run (-dummy) to see if it is going to do what we want:
combine_images.pl -newtag=combined -reduceima=2:2:reduc -pixsize=2.41 -remove -dummy
# ok, now the real thing (just leave off -dummy):
combine_images.pl -newtag=combined -reduceima=2:2:reduc -pixsize=2.41 -remove
see also:
combine_images
/storage/big1/raz/GEDMAT
mask_gedmat_images.pl
overlap_images.pl
Takes output from parsepos.pl and calculates how the images overlap so they can then be combined easily (-create option).
overlap_images.pl [options]
options:
-xdim=# xdim of image in pixels, default = 1392
-ydim=# ydim of image in pixels, default = 1040
-zdim=# zdim of image in pixels, default = 25
-pixsize=# size of pixels in um, default = 0.645
-create=file.i2i actually combine the images into one big image
-flipx flip each image around its x-axis prior to inserting it into the new image (if -create is specified)
-flipy flip each image around its y-axis prior to inserting it into the new image (if -create is specified)
-verbose
-dummy just echo what would be done, but don't do anything (nothing normally done anyway unless -create specified).
see also:
parsepos.pl
insert_image called by this program if -create is specified.
reduceima if -create is used, this may be needed to make the image small enough to see via play.
example:
parsepos.pl -pre=/storage/big3/kdb/Zeiss/20121004/LectinExp3/ -post=_1_1_20121004_LectinExp3.i2i \
/storage/big3/kdb/Zeiss/20121004/LectinExp3/LextinExp3.pos | overlap_images.pl -create=newimage.i2i
reduceima -A -bin 4 4 newimage.i2i newimage_reduced.i2i
note: you may need /storage/big1/lml/Linux/bin and /storage/big1/kef/Linux/bin in your path
source code in /storage/big1/lml/Corvera/Olga/data/ptrs
see also: insert_image
http://bigwww.epfl.ch/algorithms.html -> MosaicJ (which may already exist in Fiji)
parsepos.pl
Parse a position file produced by Zeiss system to pull out x y coords for each of the images taken in well C09. Output triples (well_id x y) written to stdout. Position may be in microns, x increasing to the right, y increasing down. parsepos.pl filename.pos options: -well=string well to analyze. default = C09 -pre=prestring add this string in front of the well_id -post=postring add this string after the well_id these two options are needed if you want to pass this output to overlap_image.pl -create=newimage.i2i, since that will need to be able to find the image associated with each well_id. -verbose additional output to stderr -vverbose additional output to stderr example: parsepos.pl -pre=/storage/big3/kdb/Zeiss/20121004/LectinExp3/ -post=_1_1_20121004_LectinExp3.i2i \ /storage/big3/kdb/Zeiss/20121004/LectinExp3/LextinExp3.pos | overlap_images.pl -create=newimage.i2i see also: overlap_images.pl insert_image note: you may need /storage/big1/lml/Linux/bin and /storage/big1/kef/Linux/bin in your path.
prep_and_restore.pl
Restores all the i2i files in the current directory. Skips any files with _.i2i or _r.i2i in their name.
Also ignores any files which do not have dat or ctx in their name.
options:
-dummy just print what would be done
-bkgnd=# pass this through to find_bkgnd as the -a option. eg, -bkgnd=-.10 will lower the bkgnd value
which is subtracted off by 10 percent.
-smooth=# smoothness factor (smaller is less smoothing). default = .00002
-iterations=# maximum number of iterations, default = 999
-scale=#:# GFP and RFP output scaling for restoration (respectively). default = 1.0 and 1.0.
-verbose echo what is being done
-maxfiles=# only analyze this many files. primarily for debugging
-twod during background subtraction, vary the background value for each 2D slice, instead of
just using the same value for all z slices.
-files=:file.i2i:...:filen.i2i just restore these files. and do it even if _.i2i or _r.i2i exists.
-dont_restore do the data prep (filename_.i2i and filename__.i2i produced), but don't do the restoration (but not filename__r.i2i).
-beowulf queue the restorations to run on our beowulf (dispatched via qsub, using pbs (torque), from a daemon running on m13)
rather than immediately on the localhost.
see also: pbsnodes (on m13, only servers running fc16 or fc17 are on the allowed list right now)
qstat -f (on m13) will show lots of info on each job.
If this is not specified, each restoration is run sequentially, with this script blocking while waiting for each one.
-gpu only use this if you are logged into zirconium. It will run the restoration on zirconium using the GPU version
of the restoration.
-outdir=dir rather than place the results in the current directory, place it in the fully qualified specified directory.
leave off the last /
-GFPpsf=file fully qualified path to GPFpsf (used when file name is *_#_#_1*.i2i, eg, file_0_1_1.i2i)
default = /storage/big1/lml/Corvera/Olga/data/GFPpsf_segxyz_.i2i
note: psf should have background values of ~0, otherwise 0-padding will produce blocking artifacts.
-RFPpsf=file fully qualified path to RPFpsf (used when file name is *_#_#_2*.i2i, eg, file_0_1_2.i2i)
default = /storage/big1/lml/Corvera/Olga/data/RFPpsf_segxyz_.i2i
note: psf should have background values of ~0, otherwise 0-padding will produce blocking artifacts.
-silent print less info
examples:
cd /storage/big3/kdb/Zeiss/20121004/LectinExp3
/home/lml/krypton/Corvera/Olga/prep_and_restore.pl -files=G11_0_1_1_20121004_LectinExp3.i2i:G03_0_1_2_20121004_LectinExp3.i2i
-beowulf -outdir=/storage/big1/lml/Corvera/Olga/data
see also:
epr_beowulf a GUI front end for submitting individual restoration jobs to the beowulf queue
/storage/big1/epr/bin/cuda/epr_gpu -cuda 1 if logged into zirconium, will run a single job on the graphics card (like -gpu option).
prepdata
preppsf
note: /storage/big1/lml/Linux/bin /storage/big1/lml/Linux/bin64 and /storage/big1/kef/Linux/bin probably need to be in your
path to successfully run this script.
prepfiles.pl
prepfiles.pl [options] Takes a dual-wavelength image and splits it into two separate images (segim_huge), applies scratchng, and prepdata. Run this from a directory. It will be applied to all images in the directory. There must be a file called dark.i2i in the directory too. Removes all intermediate files and original 2wavelength image. options: -safe : don't delete the original dual-wavelength image -echo : echo what is being done
recalc_quadrant2.pl
align_quadrants.pl calls this program. It is easier to use it than to call recalc_quadrant2.pl manually.
This program converts from pts printed by play (so in the coordinate system gotten from combining the four quadrant images of a well (with no overlap))
to one used by register_images.pl (ie, each pt is in the coordinate system of its own individual image).
It also pairs the points up and orders them the way that register_images.pl requires.
It assumes the user picked matching pairs of pts in order when using play. Eg, first a pt was picked (s key while holding down middle mouse button),
then its matching point in a neighboring quadrant was picked. This was then repeated 3 times (so 3 pairs of points are present).
note: the file names for the files associated with each quadrant are not stored in the output by default, just the quadrant number,
use -qnames to change this; they are needed if you want to subsequently have register_images.pl use chained.txt to actually combine the images.
recalc_quadrant2.pl [options] < pts.txt > chained.txt
options:
-res=# resolution the display was downsampled by prior to getting coords (ie, what was used in chres, see below).
if chres was .5, then -res=2, ie, 1/chres value. default = 2.
-xdim=# default = 1392
-ydim=# default = 1040
-qnames=name0:name1:name2:name3 file names associated with the images used to produce each of the 4 quadrants.
they must be in this order. If not specified, then the output will not be
usable by register_images.pl (unless -root is implemented there).
-outimage=image.i2i also produce an output combined image (ie, call register_images.pl, see below).
-qnames required for this option.
-blend if -outimage is specified, then blend the overlapping images to produce the output image.
-verbose print overlap info to stderr. maybe to use in -o option to combine_images.
-debug stuff to stderr
more info. the quadrants in the combined image (see combine_images below) are numbered as follows:
quadrant 1 | quadrant 3
quadrant 0 | quadrant 2
where x increases to the right, y increases up
example:
# combine the 4 well images without overlap to see what pts might match between quadrants:
combine_images -o 0 F06_0_2_0_20140505_GEDMAT_P025_WES_OM_Day14.i2i F06_1_2_0_20140505_GEDMAT_P025_WES_OM_Day14.i2i
F06_2_2_0_20140505_GEDMAT_P025_WES_OM_Day14.i2i F06_3_2_0_20140505_GEDMAT_P025_WES_OM_Day14.i2i tmp.i2i
# look at the resulting image and identify matching pts:
chres -S .5 .5 1 tmp.i2i - |play - # save (s) pairs of pts which match between the quadrants. print them using z. create file F06_pts.txt:
marked points
x= 769 y= 497 z= 1 set= 1 value= 3099
x= 799 y= 525 z= 1 set= 1 value= 3684
x= 709 y= 819 z= 1 set= 1 value= 4039
x= 682 y= 854 z= 1 set= 1 value= 3974
x= 518 y= 497 z= 1 set= 1 value= 3257
x= 543 y= 524 z= 1 set= 1 value= 3436
# now use this program to multiply by 2, subtract xdim (1392) and/or ydim (1040) as appropriate, pair and order pts.
recalc_quadrant2.pl -outimage=F06_combined.i2i -qnames=F06_0_2_0_20140505_GEDMAT_P025_WES_OM_Day14.i2i:F06_1_2_0_20140505_GEDMAT_P025_WES_OM_Day14.i2i:F06_2_2_0_20140505_GEDMAT_P025_WES_OM_Day14.i2i:F06_3_2_0_20140505_GEDMAT_P025_WES_OM_Day14.i2i < F06_pts.txt > F06_chained.txt
# same thing, but in two steps.
recalc_quadrant2.pl -qnames=F06_0_2_0_20140505_GEDMAT_P025_WES_OM_Day14.i2i:F06_1_2_0_20140505_GEDMAT_P025_WES_OM_Day14.i2i:F06_2_2_0_20140505_GEDMAT_P025_WES_OM_Day14.i2i:F06_3_2_0_20140505_GEDMAT_P025_WES_OM_Day14.i2i < F06_pts.txt > F06_chained.txt
register_images.pl -zdim=5 -xscale=1 -yscale=1 -post= -debug -out=F06_combined.i2i F06_chained.txt
see also:
/home/lml/krypton/Corvera/Raz/README 5/15/14
align_quadrants.pl
recalc_quadrant.pl
combine_images
combine_images.pl
register_images.pl
insert_image2
register_images.pl
Takes a bunch of images taken in one well on the microscope, translates them
based on ChainedTrans.txt and combines them into one big image.
ChainedTrans.txt should be a partially ordered file with entries like
X02:X03 (685,282):(736,287)
This means that location (685,282) in file X02 is the same as location (736,287) in file X03.
By partially ordered I mean that for file1:file2 file1 should already have been the
right hand side of a similar line previously (so its position is already defined relative to
the file on the left hand side), unless it is the very first line in
the file. So every right hand side is relative to a left hand side which has already
had its position defined.
Output goes to new_trans.i2i
register_images.pl [options] ChainedTrans.txt
-out=filename.i2i name of output file produced. default = newimage.i2i
-post=string the rest of the filename after what is in ChainedTrans.txt, eg, if the file is named
X01_0_0_1.i2i, -post=_0_0_1.i2i (this is also the default for post if not specified).
-xdim=# size of original each original image (before recaling by maxtrima, see example below). default = 1392.
-ydim=# default = 1040
-zdim=# default = 21
-xscale=# binning done by matrixima (see example), coords will be scaled by this. default = 4.
-yscale=#
-blend when combining images, blend the values where they overlap (default is to just replace any previous
value with the new value).
-dummy
-debug
-h
-shift no circular shift (see tranima). old.
note: tranima will pad images with -32768 which play will show in red (I think) and DAVE will be bothered by.
example:
matrixima -row 4 W X Y Z -col 3 01 02 03 -pos 0 -obj 0 -fil 1 new.i2i
play new.i2i -> ChainedTrans.txt
register_images.pl ChainedTrans.txt
dave -I new_trans.i2i -K -z 15 # a very big file.
source code: ~/krypton/bin/
see also:
combine use this on newer images which have spatial information stored in their header
parsepos.pl
~/krypton/Corvera/Olga
/storage/big1/lml/Corvera/Olga
recalc_quadrant.pl
recalc_quadrant2.pl
align_quadrants.pl the simplest manual method of aligning the four images in the well
smooth_gaps.pl
This program applies smooth_gaps to all files named *reduced.i2i and produces files named *reduced_sg.i2i (unless -tag changed it). usage: smooth_gaps.pl -run [options] options: -tag=something tag to add before the .i2i in the new file name. -verbose print out each filename as it is being smoothed. -dummy just echo what would be done, but don't do anything see also: smooth_gaps
trans.pl
Takes a bunch of images taken in one well on the microscope, translates them
based on ChainedTrans.txt and combines them into one big image.
ChainedTrans.txt should be a partially ordered file with entries like
X02:X03 (685,282):(736,287)
This means that location (685,282) in file X02 is the same as location (736,287) in file X03.
By partially ordered I mean that for file1:file2 file1 should already have been the
right hand side of a similar line previously (so its position is already defined relative to
the file on the left hand side), unless it is the very first line in
the file. So every right hand side is relative to a left hand side which has already
had its position defined.
Output goes to new_trans.i2i
register_images.pl [options] ChainedTrans.txt
-out=filename.i2i name of output file produced. default = newimage.i2i
-post=string the rest of the filename after what is in ChainedTrans.txt, eg, if the file is named
X01_0_0_1.i2i, -post=_0_0_1.i2i (this is also the default for post if not specified).
-xdim=# size of original each original image (before recaling by maxtrima, see example below). default = 1392.
-ydim=# default = 1040
-zdim=# default = 21
-xscale=# binning done by matrixima (see example), coords will be scaled by this. default = 4.
-yscale=#
-blend when combining images, blend the values where they overlap (default is to just replace any previous
value with the new value).
-dummy
-debug
-h
-shift no circular shift (see tranima). old.
note: tranima will pad images with -32768 which play will show in red (I think) and DAVE will be bothered by.
example:
matrixima -row 4 W X Y Z -col 3 01 02 03 -pos 0 -obj 0 -fil 1 new.i2i
play new.i2i -> ChainedTrans.txt
register_images.pl ChainedTrans.txt
dave -I new_trans.i2i -K -z 15 # a very big file.
source code: ~/krypton/bin/
see also:
combine use this on newer images which have spatial information stored in their header
parsepos.pl
~/krypton/Corvera/Olga
/storage/big1/lml/Corvera/Olga
recalc_quadrant.pl
recalc_quadrant2.pl
align_quadrants.pl the simplest manual method of aligning the four images in the well
blur3d
Blurs an image by a psf.
FFTs are done with 4 byte floats, so total space needed will be 4 times the padded image size (in voxels)
plus the size of the two original images (image and psf)
The input image can be real or short int format. The psf must be short int format.
note: performs a convolution (not a correlation).
note: fixed a small bug. 10/17/11
Usage: blur3d [options] image psf newimage
options:
-S #: scale (multiply) output values by # (to prevent underflow or overflow)
voxels which after scaling are > 32767 are set to 32767. voxels < -32766 are set to -32766.
-p # # #: pad the image in x, y, and z by the specified amounts
by default image is padded (with 0) by 1/4 psf width all around,
so its size for blurring becomes bigger by psf/2.
psf is always padded to the same size the image becomes.
Padding is stripped off before newimage is written, so it will be the same size as
the original image.
-N # # #: pad so that the image has the specified dimensions after padding (just prior to FFT).
-n #: pad with # instead of boundary pixel values.
-Z: pad with zero instead of the boundary pixel values.
-c # # #: the center of the psf is at (#,#,#) zero indexed.
the default is to take the brightest pixel as its center.
-C: don't center the psf at all (default is to center on the max, unless -c is specified).
-P: normalize the psf so each slice (after padding) averages to 1. -P acts like an optical PSF.
the default is to normalize so the entire psf (after padding) has an average pixel value of 1
(the default keeps total light constant).
-d: don't normalize at all.
-o: do a cross correlation instead of a blur (convolution)
-R: output image as 4 byte floating point instead of short integer.
-v: verbose. print more info to stderr.
note: entire image name (with extension) is NOT required.
note: can read from stdin or write to stdout (use a dash instead of the file name)
note: a compressed image will be automatically be uncompressed.
Copyright 2010 University of Massachusetts Medical School and the Biomedical Imaging Group
All rights reserved. Explicit permission to use this program must be received prior to use.
canny
This program reads in a 2 or 3d image and outputs the results of a 2D Canny edge detector on it. Canny is the derivative (in the gradient direction) of the magnitude of the first derivative (so it is a type of 2nd derivative). NOTE: see option -n below for an exact answer. Each z slice is treated independently of the others. Its application will often be preceded by a Gaussian blur and followed by the calculation of zero crossings. If an INT type image is input, an INT is output. If a REAL type image is input, a REAL is output. Usage: canny [options] infile outfile options: -d: Debugging flag -n: normalize the result. This option causes the true 2nd derivative to be calculated. Without it, only the sign of the second derivative is guaranteed to be correct (which is sufficient if only zero crossings will be calculated from it). Divides each point by Ix*Ix+Iy*Iy. -s #: scale factor to multiply all output values by. This is useful if you are outputing an INTEGER image with a very small range of values which would otherwise all be truncated to just a few numbers. note: entire image name (with extension) is NOT required. note: a - in place of an image name means stdin or stdout. Example usage: To create a gradient edge strength image for a shrink wrap run: blur_spatial image.i2i -|canny - -|zerosu - - |blur_spatial - out.i2i
cblur
Usage: cblur [options] arg1.i2i arg2.i2i For every zero valued point in each xz plane, cblur replaces the zero value with the -100 times the distance from the nearest non-zero voxel in the xz plane. Options: None at this time. Note: entire image name (with extension) is required. Note: a - in place of an image name means stdin or stdout.
center_of_mass
This program reads in a 4D image and translates each 3D image so its center of mass is moved to be the center of the image. One 3D image is read at a time. Usage: center_of_mass [options] inimage.i2i outimage.i2i options: -t tdim: number of time points, default = 1. This is NOT read from the header. -T #: threshold inimage.i2i prior to analysis. voxels <= # ignored, default = 0 -v: verbose note: source code in krypton:/home/lml/krypton/facil directory.
combine_images
This program reads a bunch of image files which have their xy position within a well stored in their header. It then combines the images. Karls acquisition program for the Zeiss microscope should automatically add this information to the header. A maximum of 50 images can be combined. Images must all be the same size. Images are also assumed to be aligned in z already. Images are added to the output image in the order specified on the command line, with later pixel values replacing previous values if there is an overlap of the images. Usage: combine_images [options] im1.i2i im2.i2i [more input images] outim.i2i options: -d debug. stuff to stderr. -f do NOT fill pixel locations which have no data with nearest data value (added 2/3/14) Filling is done by default to try to avoid introducing sharp intensity edges into the image. -X flip each input image in X before using it (-Y can also be specified at the same time) -Y flip each input image in Y before using it -p # over-ride the specified pixel size with this one (in um). The 2.5X objective has a pixel size of 2.58 um stored in the image header (for images acquired prior to 4/13/13) but it should be 2.41 -x #:#[:#...] rather than use the x positions stored in the image header use these x pixel positions. these should be the x offset of one image relative to another, in the same order as the image names on the command line (there MUST be one number for each image). For example: combine_images -x=0:500:0:500 im11.i2i im21.i2i im12.i2i im22.i2i outim.i2i if im21.i2i and im22.i2i are displaced 500 pixels to the right of the other two images. -y #:#[:#...] same, but for displacement in the y direction. Since these are pixels, they should be integers. note: many times images are 1392x1040 and overlap by 50 pixels, and image 0 is bottom left image 1 is top left, image 2 is bottom right, and image3 is top right, then: -x 0:0:1342:1342 -y 0:990:0:990 should work -o # rather than specifying -x and -y, thus just specifies the overlap (e.g., 50) and figures out the rest for you (assumes images ordered BL,TL,BR,TP - as above - and x and y overlap the same works for any size image. note: entire image name (with extension) is NOT required. note: a - in place of an image name means stdin or stdout. note: compressed images are temporarily uncompressed into /usr/tmp. See also: parsepos.pl (not needed now that Karl fills the image header with this info) combine_images.pl applies combine_images (and, optionally, reduceima) to all files in a directory. register_images.pl align_well_images.pl pre-processor for combine_images if the y-coord in the header is invalid. insert_image Source code in ~/krypton/facil/combine_images.c combine_images:
flood3dL
Usage:
flood3d [options] input.i2i output.i2i
Description:
flood3d reads in an image and outputs an image with each region's
voxels all at the same intensity (intensity = 1 for 1st region, 2 for 2nd
region, etc.). Regions are 26 connected voxels greater than the threshold.
If no output image is specified no output image will be produced.
Options:
-t #: Value to threshold image (default 0).
-s #: Value to start numbering objects (default 1).
-r x y z: Mark the region at x y z (1 indexed).
this option can be specified up to 5 times.
If z < 0, then starting points x y 1, x y 2, .. x y zdim
are used. Use z < 0 in conjunction with -c 4 or -c 8.
all such regions found are marked with the same region number.
-c #: Do # connected flood fill (default 26).
Options are 4, 8, or 26 connected flood fills.
4 or 8 connected flood fills are for 2D flood fills
with the fill being done in x, y for each z plane.
(each slice will be done, unless -r is specified).
-m #: only keep regions whose size in voxels is > #. Default = 0.
-b: Output will be binary (regions counted will be 1,
regions not counted will be 0.
-q: Quite mode, fewer messages printed.
Caveats:
Threshold and number of objects has to be less than 32,000.
Note:
Entire image name (with extension) is required.
A - in place of an image name means stdin or stdout.
Keywords:
count objects, flood fill
Additional programs:
patchstat do_transform
gauss_deriv
This program takes an image, blurs it with a Gaussian of specified standard deviation and then takes derivatives of the image It does all of this in the frequency domain. Usage: gauss_deriv [options] infile.i2i outfile.i2i options: -a angle: angle of rotation (in radians) for the x,y axes about z. default = 0.000000 -x order: number of derivatives in the x direction. default = 0 -y order: number of derivatives in the y direction. default = 0 -p x y z: pixel size in microns (zsize ignored for now), default = (0.1,0.1,0.1) -s dev: standard deviation in spatial domain of Gaussian (in microns). -S dev: standard deviation in frequency domain of Gaussian (cycles/um). default = 1.000000 -e epsilon: cutoff for Gaussian intensity calculation. default = 0.040000 -r newmax: rescale output 0-newmax. newval = (oldval-min)*newmax/(max-min). This is useful especially when all the values would otherwise be < 1. This also prints to stderr the min and max values before rescaling. -R scale black: rescale output: newval = (oldval-black)/scale . -v: verbose -n: no op. just take a forward and inverse transform. For debugging. note: the entire image name is NOT required. note: compressed images will be uncompressed. note: a - in place of an image name means stdin or stdout. restrictions: only works in 2D. 3D images have each z plane processed separately. see also: gauss_deriv_unc, blur_spatial, canny, zerosu.
i2i2mhd
This program just creates a MetaImage header file (file.mhd) which can be read, in conjunction with the original
i2i file, into a ITK or VTK program. NOTE: you must keep inimage.i2i and inimage.mhd together in the same directory.
If specified with a .mha extension then the resulting image has both the ascii header and the data together
in one file (not done yet).
Usage: i2i2mhd inimage.i2i inimage.mhd
i2i2mhd inimage.i2i inimage.mha
options:
-t # : set output tdim to the specified number. specify 0 if the image has no time dimension.
-l : force output to be designated little endian
-b : force output to be designated big endian
-s dx dy dz : pixel spacing. Some of the ACME routines may look at these values.
Examples:
i2i2mhd test.i2i test.mhd
~/krypton/segmentation/MR-Source/ACME/ACME-Binary/bin/cellPreprocess test.mhd test_proproc.mhd .3
the above will create test_preproc.zraw and test_preproc.mhd, if, instead, you specify
test_resampled.mha as ouput to cellPreprocess you get all the output in the mha file.
See also: mhd2i2i
http://www.itk.org/Wiki/MetaIO/Documentation, ~/krypton/ITK/README.lml.build-4.3,
~/krypton/segmenation/MR-Source/README.lml, /home/lml/krypton/segmentation/MR-Source/ACME/lmltest/README
source code in: /home/lml/krypton/facil
insert_image2
incorrect command line, too few args.
insert_image2 does a weighted merge of pixels in an original image with overlapping pixels from replacement images.
Pixels which fall outside the original volume will be ignored.
WARNING: the original image is MODIFIED, a NEW image is NOT produced (unless -n is specified).
The weighting function for the blending makes the most sense for kind of usage align_quadrants.pl makes of it;
ie, blending four quadrant images which only overlap a little and are being inserted into an initial blank image.
Usage: insert_image2 [options] original_image.i2i [replacement_part.i2i xpos ypos zpos]
replacement_part.i2i xpos ypos zpos: the replacement image and the x,y,z coordinate (0-indexed)
that it should start at. This option can be repeated as many times as desired.
options:
-r # if a pixel in original_image.i2i has this value, REPLACE it with any overlapping pixel (not a weighted avg).
note: it could still then be modified by subsequent replacement images (they are analyzed in command line order)
-R # if a pixel in a replacement image has this value, use it to REPLACE any overlapped pixel (not a weighted avg).
note: it could still then be modified by subsequent replacement images (they are analyzed in command line order)
-k # if a pixel in th original_image.i2i has this value, do NOT replace it.
-v # same as -k (for backwards compatibility). keep this value.
-n newim.i2i rather than over-write the original image, create a new image with this name.
-d debug. some info gets printed to stderr
example:
parsepos.pl -pre=/storage/big3/kdb/Zeiss/20121004/LectinExp3/ -post=_1_1_20121004_LectinExp3.i2i \
/storage/big3/kdb/Zeiss/20121004/LectinExp3/LextinExp3.pos | overlap_images.pl -create=newimage.i2i
see also: /storage/big1/lml/Corvera/Olga/data/ptrs/README
insert_image -the same as insert_image2, but doesn't perform any weighted merging.
smooth_gaps
combine
combine_images.pl
align_quadrants.pl which calls combine_images and recalc_quadrant2.pl which calls register_images.pl which calls insert_image2
(or insert_image if -noblend was passed to align_quadrants.pl).
source code in: /home/lml/krypton/facil
mergeimL
Merge a bunch of 2 D files of same dimensions. The first image can be a 3 D image. If -x and -y are specified then all images are padded (on high x and high y sides) to the same specified dimensions, so the initial images can be different sizes. This version can read shorts or floats (but all must be the same) Usage: mergeimL [options] im1 ... imN newimage.i2i options: -r : remove original files as they are merged. note: if the program bombs for some reason during this, data will be lost. -b: merge the images in backwards order from the list (ie, the last image will be the first slice). Use with -b in extractslice. -x xdim: pad all images to this x dimension. -y ydim: pad all images to this y dimension. NOTE: this permits images of different sizes to be merged by padding to the same size. -p #: when padding (-x and -y options) use this value (0 default) -d: print out some debugging information. note: entire image name (with extension) is required. note: source in krypton:/home/lml/krypton/facil
mhd2i2i
This program reads a MetaImage file (file.mhd or file.mha) and converts (casts) it into a (short int) i2i image file.
Usage: mhd2i2i inimage.mhd outimage.i2i
options:
-p : just print some info about the image and quit. outimage.i2i does not need to be specified in this case.
-s # # : multiply values by the first number and add the second, then cast to a short int.
-d debug
Examples:
mhd2i2i test.mhd test.i2i
See also: i2i2mhd, http://www.itk.org/Wiki/MetaIO/Documentation, ~/krypton/ITK/README.lml.build-4.3,
~/krypton/segmenation/MR-Source/README.lml, /home/lml/krypton/segmentation/MR-Source/ACME/lmltest/README
mayavi2 (which can view .mhd files directly)
source code in: /home/lml/krypton/facil
oib2i2i
oib2i2i takes an Olympus oib file and converts it into one or three i2i files.
note: it should also work with many other file types, including DeltaVision files.
java oib2i2i [options] file.oib file.i2i [z dimension will be the number of channels]
java oib2i2i [options] file.oib file_red.i2i file_green.i2i file_blue.i2i
java oib2i2i [options] file.oib file_red.i2i file_green.i2i file_blue.i2i file_alpha.i2i
options:
-l[ittle]: force interpretation of 2 byte pixel values to be little endian (I may have this backwards)
-b[ig]: force interpretation of 2 byte pixel values to be big endian (I may have this backwards)
-v[erbose]: verbose. prints info about file contents as planes are converted.
-d[ebug]: prints debug info to stderr.
-i[nfo]: like verbose. but ONLY prints info about file contents, then stops
must still give it a (dummy) output file argument
see also:
GetMicroscopeImageInfo file.oib :will produce lots of information about the file
http://www.loci.wisc.edu/software/bio-formats
source in /home/lml/krypton/packages/BioFormats/oib2i2i.java
entered main()
pwarpL
pwarp warps an image into a new image based upon a set of
registered points from one image to another. That is, the
transformation necessary to transform one set of registered
points into the other is calculated. Then this transformation
is applied to the image specified, producing a new image.
A line segment can also be specified as an old "pt". The new
pt will match the best position along this line segment.
The pointfile should contain points in the following format,
new x and y dimensions, number of pts, new pts, oldpts. for
example if 10 pts are given:
300 240
10 points
new_x1 new_y1
. .
. .
new_x10 new_y10
initial_x1 initial_y1
. .
. .
L old_lowx old_lowy old_hix old_hiy (this is a line segment)
. .
. .
initial_x10 initial_y10
the points can be in floating point.
Usage: pwarp [options] oldimage pointfile newimage
options:
none.
Note: the .i2i extension is NOT necessary.
rawtoi2i2
Converts a "raw" image into our i2i format. The raw image can be alternating red, green blue values or either 1 byte per color channel or 2 bytes per color channel. It can also be grayscale (-n 1) and 1,2, or 4 bytes per channel. Usage: rawtoi2i [options] -d xdim ydim zdim inimage.rgb outimage.i2i Options: -d xdim ydim zdim: dimensions of inimage.rgb. This MUST be specified. -b #: 1 or 2 bytes per color channel, default = 1 if the image is grayscale (-n 1) this can be 4 -c #: which color channel to use (0=r, 1=g, 2=b), default = 0 ignored if grayscale (-n 1). -n #: 3 if color channels (rgb), or 1 for grayscale. -S scale black: newval = (oldval-black)/scale. -m min max: after rescaling, if newval < min, newval = min. defaults = -32765,32765 -s: values in inimage are signed values (not unsigned). -e: swap bytes (change endian). only make sense if -b 2 or 4 -v: verbose Examples: identify inimage.tif :get image dimensions (xdim,ydim,zdim) /usr/bin/convert inimage.tif -channel Red inimage.rgb rawtoi2i -d xdim ydim zdim inimage.rgb outimage.i2i See Also: cstoi2i, zeisstoi2i, convert (man ImageMagick), imformat, imformatb. Bugs: the created image has a strange permission. fix it by doing: chmod 644 outimage.i2i Source code in ~krypton/lml/facil
replace_slicesL
This program reads a 3D image and replaces the specified slices with a weighted composite of the closest surrounding slices which are not being replaced. It is useful for getting rid of bad slices from an image. The weights perform a linear interpolation. Usage: replace_slices [options] inimage.i2i outimage.i2i options: -r # s1 s2 s3, etc : replace # slices (e,g,. 3 slices); the slices to replace are s1, s2, s3, etc. NOTE: the slices must be sorted (s1<s2<s3). NOTE: slices are 0-indexed (first slice is 0). NOTE: the first and last slices in the image cannot be replaced. note: entire image name (with extension) is NOT required. note: a - in place of an image name means stdin or stdout. note: a compressed image will be automatically be uncompressed.
rescale
This program linearly rescales image intensities to a new intensity range: 0 (new_min) to 1023 (new_max) by default. Usage: rescale [options] inputimage outputimage options: -N #: New max should be #. -n #: New min should be #. -M #1 #2: any original pixel value greater than # should be set to #2 before any rescaling takes place. -m #1 #2: any original pixel value less than #1 should be set to #2 before any rescaling takes place. So if you have a few munged values, ie, large negative numbers which you don't want to mess up the rescaling you can just use -m 0 0 on the command line. -v old new: voxels with value old will just be set to value new. note: "old" cannot be within the range which other pixels will be rescaled to. -b write out an i2i byte image (rather than a short). values > 255 will be truncated. note: entire image name (with extension) is NOT required. note: a - in place of an image name means stdin or stdout. note: compressed images are temporarily uncompressed into /usr/tmp.
Miscellaneous
analyze_cross_sections_of_voronoi.pl
This program reads files produced by cross_sections_of_voronoi (-P)
Results prints volume (x) vs predicted volume (y) to stdout.
analyze_cross_sections_of_voronoi.pl [options] file1.txt [... filen.txt]
options:
-debug info about all fields on each line printed to stderr
-stdout everything to stdout
-print_avg for each file, just graph mean and std error of the mean, rather than all data pts
examples:
cross_sections_of_voronoi -P voronoi.i2i > filename1.txt
cross_sections_of_voronoi -P voronoi.i2i > filename2.txt
axnalyze_cross_sections_of_voronoi.pl *.txt
see also:
/storage/big1/lml/Raz/voronoi/README, true_vs_predicted.xmgr
/home/lml/krypton/Corvera/Raz/stereology/README, run_voronoi.pl, run_voronoi2.pl, compare_volume_measurements.pl, plot_results.pl
cross_sections_of_voronoi
/home/lml/krypton/packages/fiji/Fiji.app/macros/stereology_lml.txt
/home/lml/krypton/packages/fiji/run_stereology.txt
/storage/big1/lml/Raz/voronoi/
neartrack.pl
neartrack.pl
find which track a specified coordinate is near (ie, with dx,dy) (eg, DAVE crosshairs gives us a coord, what track is it?)
assumes coords passed are zero indexed
usage: neartrack.pl -x=# -y=# [-z=#] [-dx=#] [-dy=#] [-oneindexed] file.rpts
parse_macros4.pl
Can't use 'defined(%hash)' (Maybe you should just omit the defined()?) at /home/lml/krypton/bin/parse_macros4.pl line 562.
path_diff.pl
compare the differences between the paths found from two different runs of exo5 path_diff.pl file1 file2 options: -dx=# x coord at fusion time can differ by this and still be considered same path (default = 0) -dy=# y coord at fusion time can differ by this and still be considered same path (default = 0) -df=# fusion time can differ by this and still be considered same path (default = 0) -debug -Debug
trackinfo.pl
# analyze output from "track or track -r".
# assumes coords passed are zero indexed.
# usage: trackinfo.pl -x=# -y=# [options] file.rpts
# options:
# -reg=# : just print info about the specified region number
# -track=# : just print info about the specified track
# -trackcoords=minsize:maxsize:miniod:maxiod:minlife:maxlife
# : print the coords (rpts format) of all tracks which satisfy
# the criteria (or all its regions satisfy them). lifetime is # of timepts it exists for.
# -tdist=mindist:maxdist: use with -trackcoords to specify min and max (xy manhattan) distance track travels
# -micro: print coords of all paths. color code them so if they connect at all
# they are the same color (eg, all along the same microtubule)
# -x=# -y=# : find which track the specified coordinate is near (eg, DAVE
# crosshairs gives us a coord, what track is it?)
# -z=# : also include z in distance calculation.
# -dx=# : dx, default = 3
# -dy=# : dy, default = 3
# -dz=# : dy, default = 1000 (ie, essentially ignored)
# -oneindexed : otherwise -x, -y, -z assumed to be zero indexed (rpts file is always one indexed)
# -t : print additional region and track info (which should be in file.rpts
# if -r was specified when running "track") for objects found
# (eg, via -x=#,-y=#,or -region=#)
# -sf=name : sort field. name of field to sort on when printing out a track.
# implies -t. default = time ("set");
# -rs : reverse the "sense" of the sort (normally smallest to largest). implies -t.
# -pf=field1:field2:field3 : print fields. which fields to print from the region info (implies -t).
# default is all fields. you can put as many fields as you want here,
# separated by colons.
# -verbose: : print info about progress to stderr
# example:
# track -a 3 -n 400 -r -s 0 5 -S -t 25 -v 5 500 -d 15 9 -u .33 .33 .25 5 -h .01 a.vel a.i2i > a.rpts
# trackinfo.pl -x=52 -y=28 -t -pf=set:size:nextid a.rpts
# trackinfo.pl -reg=103 -t -pf=set:size:nextid a.rpts
find_bkgnd
find_bkgnd takes a 3D image and tries to estimate a background intensity value. It does this by examing the IOD in each of the four corners of the image, picking the average pixel value associated with the lowest IOD. IOD is calculated in a square region whose width (default = 2) can be changed with -r. The IOD is the total light in this square region added across all z-planes. The avg pixel value is printed to stdout. Usage: find_bkgnd image1 options: -r #: change width of region examined, default = 2 -c also print out the x and y coord (1-indexed) of the corner producing the avg value. -a # add this fraction to the value. eg: -a -.10 will subtract 10 percent from whatever avg value would have otherwise been printed out, so we don't subtract quite so aggressively. see also: ~/krypton/Melikian/find_and_restore.pl source code in: /home/lml/krypton/facil
interp
interp takes a list of contours from an rpts file and
interpolates new contours between them at each zslice that
was skipped.
METHOD: By default it does this by taking the fft of
each curve, thereby obtaining fourier coefficients. These
coefficients are then interpolated between the known contours
to get coefficients for the in-between slices, then an inverse
fft is taken to produce the curve for the in-between slices.
Usage:
interp [Options] input.rpts output.pts
Options:
-f #: number of positive frequency terms to use,
default = 9
-l: perform linear instead of cubic spline interpolation
on the parameters describing the existing outlines.
-o name: only read object "name" from the rpts file.
-n #: number of pts to resample each slice.
(all #'s factors must be 2)
-S: do interpolation in spatial domain. do not take fft.
this is only a linear spatial interpolation.
-a: angularly interpolate positions (fft not used).
this is used if we are interpolating curves, such
as microtubules. The beginning of the curve is linearly
interpolated (the mtoc location), all other points have
their distance and angle from the mtoc linearly interpolated
between their initial distance/angle and final distance/angle.
NOTE: the angular change must by < 180 between defined curves.
NOTE: only one open curve is allowed per image slice.
-p #: pixel value to be output to rpts file, -3 default.
-c: add 1 more point to each contour to ensure it is closed, ie
that the first point equals the last point. (This is not necessary,
and ill-advised, for use with min_cylinder or shrink_wrap.)
NOTE: zslices must be in ascending order in rpts file.
NOTE: contours must all be drawn in the same direction
(i.e., all clockwise or all counter-clockwise).
NOTE: the name of each object is changed to "nuke" if -o is not specified.
NOTE: a maximum of 3000 zslices is allowed
see also: interp_many
source code was in: /storage/big1/lml/was_vision/interp
source code now in: /home/lml/krypton/was_big1/was_vision/interp
object_overlap
#/home/lml/krypton/bin/object_overlap
Takes images produced by countobjs -Q option or vcspat-models -n option and calculates
how many object from each image overlap
with objects from the other image. Writes to stdout.
Usage: object_overlap [options] image1.i2i image2.i2i > overlap.txt
options:
none yet.
Example:
countobjs -Q im1_obj.i2i im1.count im1.i2i
countobjs -Q im2_obj.i2i im2.count im2.i2i
object_overlap im1_obj.i2i im2_obj.i2i > im1_im2.overlap
See also: vcspat-models, vcspat, krypton/Qiong/run1
Source code in krypton/facil
Data Reformatting
chres_rpts.pl
Reads in some rpts files and rescales their x and y coords so that they will match an image which
has been similarly rescaled.
usage:
chres_rpts.pl [options] infile.rpts > outfile.rpts
options:
-sx=# rescale x -> x*#
-sy=# rescale y -> y*#
-truncate after rescaling, truncate to an integer (newer versions of addlines can handle floating pt)
-round after rescaling, round to an integer
-max=#:# don't let x become > than #1. don't let y become > #2.
-min=#:# don't let x become < than #1. don't let y become < #2.
example:
# suppose you have a floating pt rpts file
find_peaks_inrpts.pl -rpts=QS_H10+1Q_3T3_center_2_peaks.rpts:.15:.033
Results/mat_quad/QS_H10+1Q_3T3_center_2_control_q_kept.rpts > /dev/null
# and you want to see it in better spatial detail:
chres -S 4 4 1 Data/QS/QS_H10+1Q_3T3__center_2_ra7.i2i QS_H10+1Q_3T3__center_2_ra7_ch441.i2i
chres_rpts.pl -sx=4 -sy=4 QS_H10+1Q_3T3_center_2_peaks.rpts > QS_H10+1Q_3T3_center_2_peaks_ch441.rpts
addlines2 QS_H10+1Q_3T3__center_2_ra7_ch441.i2i QS_H10+1Q_3T3_center_2_peaks_ch441.rpts - |playc -
see also:
filter_rptsinz.pl
combine_Raz_and_lml_data.pl
combine data pts from Raz with my data points. results printed to stdout
usage:
combine_Raz_and_lml_data.pl [options] file1 file2
where file1 is find_vesselness_results2.txt (or something else in that format, ie with internal filenames formatted that way)
file2 is raz_results.out (or something else in that format)
options:
-cond1='xxx' a perl expression to be evaluated to determine which rows of data to analyze. default is all rows.
if it evaluates to > 0 the row will be used.
e.g., -cond='if ($tissue eq "OM") {1;} else {0;}'
e.g., -cond='if ($date == 7) {1;} else {0;}'
note the use of single (not double) quotes to protect this from the shell.
If this is not specified all data lines will produce an x and y value.
Some simple perl info:
use eq (equal) and ne (not equal) for comparing strings (which are in double quotes).
use == != > >= <= for numeric comparisons. && (logical AND), || (logical OR).
put $ in front of all variables (string variables and numeric variables).
statements are separated by semicolons. Blocks specified with braces.
if ( ) { } else { } where the "else {}" is optional. But don't forget the braces.
-unique_header1=something string which uniquely exists on the header line of file1 (so it can be identified)
-data_field1=something which column has the actual data we care about and will graph. default = rate
-data_factor1=# multiply data_field value in file 1 by data_factor prior to analyzing (or printing) it.
-group_by1=something how to match the data, eg, a datapoint in file1 will by matched to a datapt in file2 by this criteria
-cond2 same but for file2, defaults to -cond1
-unique_header2=something same but for file2. defaults to -unique_header1
-data_field2 same but for file2, defaults to -data_field1
-data_factor2 same but for file2, defaults to 1
-group_by2 same but for file2, defaults to -group_by1
-silent don't print warning to stderr when a data pt in one file does not have a match in the other file.
-debug stuff to stderr
-Debug other stuff to stdout
example:
get_find_vesselness_results.pl -gdm=014:019 -t2dm=005:020:021 > find_vesselness_results2.txt
get_dir_results.pl -verbose > raz_results.out
combine_Raz_and_lml_data.pl -data_factor1=2 -unique_header1=filename -data_field1=pixels_in_mask -group_by1=filename
-data_field2=mass -group_by2=imagename find_vesselness_results2.txt raz_results.out > Raz_lml_combined.txt
see also:
combine_with_Raz_curated_data.pl
compare_data_points.pl
boxandwhisker2.pl
xmgrace --help
http://plasma-gate.weizmann.ac.il/Grace/doc/UsersGuide.html
/home/lml/krypton/Corvera/Raz/gedmat_dummyhealthr.data
/home/lml/krypton/Corvera/Raz/README 4/2/14
gedmat_to_xy.pl used to produce gedmat_dummyhealthr.data. can also pull out arbitrary rows and columns
coords2pts.pl
Converts from coords format produced by track_pts to pts format needed by dave. Only handles output from track_pts with one object in it right now. coords2pts.pl [options] <in.coords > out.pts options: -one: add 1 to x,y,z coords (e.g, to go from zero indexed to the one-indexed coords dave expects) -name=trackname the name of the object created. dave wants different objects to have different names. -w=# for the w coord (intensity) just use the specified number (dave has problems if the number > 255) track_pts usually puts the max intensity in the w field. Example usage: track_pts -tdim 30 green.i2i green_in.coords green_out.coords track_pts -tdim 30 red.i2i red_in.coords red_out.coords coords2pts.pl -w=1 -one -name=green_track < green_out.coords > green_out.pts coords2pts.pl -w=1 -one -name=red_track < red_out.coords > red_out.pts concateima green.i2i red.i2i both.i2i dave -n 0 -u -O -z 1 -4 -I both.i2i green_out.pts red_out.pts
crosshair2bestpath.pl
Convert a list of pts produced inside DAVE by using the crosshair and the
"clear and save stack to file" option, to commandline arguments for
BestPath3D. May still need to add -t and -f options manually.
Must also change IMAGE.i2i and IMAGE.rpts args in the output.
crosshair2bestpath.pl [options] crosshair.dat > commandline.txt
options:
-image=# image name (instead of IMAGE.i2i) written out.
-rpts=# rpts name written out (instead of IMAGE.rpts)
-t=# add -t # to options written out written out for BestPath3D
-f=#:# add -f # # to options written out for BestPath3D
dup_rpts.pl
Takes an input file with one z slice per object, and, for each object named on the command line, duplicates that slice for a range of z slices. rpts file must be in xy (not xyz) format dup_rpts.pl -name1=#-# [-name2=#-# ...] < in.rpts > out.rpts or dup_rpts.pl -name1=#-# [-name2=#-# ...] in1.rpts [in2.rpts ...] > out.rpts example: dup_rptsinterp.pl -foo=1-5 -bar=2-8 < in.rpts > out.rpts This will take the zslice associated with the object named foo and duplicate it into zslices 1 through 5, and duplicate object bar into zslices 2-8.
extract_overlap.pl
Reformats data from the overlap data produced by closest_object for graphing by xmgrace. extract_overlap.pl [options] *.patchdist2 | xmgrace -source stdin -mstring=im1_im2: : pull out data for fraction of each object in image1 which overlaps with image2 object, default = im2_im1 You can also specify im1_im1 and im2_im2, in which case overlap is always 0 and the distance (-distance option) is the distance to the nearest other object in the same image. -distance : pull out distance info instead of overlap info -size : pull out size info instead of overlap info -showhist : print histogram of overlaps (or distances or sizes) instead of just a list of all of them -normmax : normalize the histogram to have a max of 1 (sets -showhist) -normsum : normalize the histogram to have a sum of 1 (sets -showhist) -normint : normalize the histogram to have an integral (area under curve) of 1 (sets -showhist) this lets xmgr->data transformations->integrate produce a cumlative prob distribution which goes to 1 Not quite correct yet. -cum : print cumulative histogram. also use -normsum if you want this to be a cdf (approaches 1 as a max). -binsize=# : binsize for the histogram (sets -showhist), default = 1 -verbose
filter_rptsinz.pl
Takes simple rpts file and only keeps certain z valued pts (they get renumbered). This is set up so that if you use segment -Z # # # image.i2i, and filter_rptsinz.pl -z=#:#:# where the numbers match in the two commands, then the resulting rpts will match the i2i image. rpts which are not in the correct slices are not kept. usage: filter_rptsinz.pl -z=#:#:# infile.rpts > outfile.rpts options: -z=#:#:# Z axis start, end and increment in pixels (1-indexed). -float keep (or convert) x,y,z values as floats. Otherwise it tries to auto-detect when to output floats. only works on rpts files which have a z value (e.g. xyz or xyzw format). see also: chres_rpts.pl
gedmat_to_xy.pl
Takes files of data produced by mask_gedmat_images.pl (either via stdin or as command line arguments, [filenames])
and writes x y columns of processed data to stdout (but -unique_header=none will allow it to work on other formats).
These files have a line with column names, and can have comment lines (start with #) or blank lines in them.
All other lines should just be columns of ascii text data. strings are allowed (not quoted, and no spaces in them).
The column header line (which can appear on any line and multiple times) must have a word (ie column name) unique
to it (since the data can also have strings in them). By default this word is assumed to be "filename"
(-unique_header can change this).
-cond (or -form if I bother to get that working) specifies a condition which decides which lines
of data to analyze. Any expression which evaluates to something > 0 will be analyzed.
-xcoord and -ycoord are the formula to use to determine what will go in the x or y column (if the -cond test is passed).
usage:
gedmat_to_xy.pl [-form='xxx' | -cond='xxx'] -xcoord='xxx' -ycoord='xxx' [options] [filenames]
xxx should contain the words (as variables) which are the column names in the output produced by mask_gedmat_images.pl.
required:
-cond='xxx' a perl expression to be evaluated to determine whether to print x and y derived from each row of data.
if it evaluates to > 0 the row will be used.
e.g., -cond='if ($date eq "date_sure") {1;} else {0;}'
e.g., -cond='if ($area > 10) {$area+5;} else {$dist/2;}'
note the use of single (not double) quotes to protect this from the shell.
If this is not specified all data lines will produce an x and y value.
Some simple perl info:
use eq (equal) and ne (not equal) for comparing strings (which are in double quotes).
use == != > >= <= for numeric comparisons. && (logical AND), || (logical OR).
put $ in front of all variables (string variables and numeric variables).
statements are separated by semicolons. Blocks specified with braces.
if ( ) { } else { } where the "else {}" is optional. But don't forget the braces.
note: you can also use variables $exarea and $exdist to refer to the area and dist fields
from the day 0 datalines. These values correspond to the explant area and its distance
from the image (ie, well) center for the same patient, well, and tissue type as the
current line being processed. Suppose you want to look only at data from day 11 if
the explant area (ie, day 0 area) was larger than 200,000 pixels:
-cond='if (($exarea > 200000) && ($day == 11)) {1;} else {0;}'
-xcoord='xxx' like -cond, but used to specify what the xcoord value should be
e.g., -xcoord='$farea/$area'; do not specify this if you specify -col#
-xcoord='$numprinted' will print 1, 2, 3, etc (ie, a count of how many datalines have been printed so far) for x.
-ycoord='xxx' like -cond, but used to specify what the ycoord value should be,
e.g., -ycoord='$dist'; do not specify this if you specify -col#
-ycoord='$numprinted' will print a count of how many datalines have been printed so far for y.
options describing input:
-unique_header=something rather than look for "filename" to determine which line contains header names, look for something
-unique_header=none if you specify the word "none" then column names will be col1, col2, ... and no header lines
should exist
options describing output:
-comment include a comment line up front showing the command line used to generate this data.
-col#='xxx' rather than just two columns, you can specify what to print in up to 20 by repeating this command
# is the column number (starting with 1). do not specify -xcoord or -ycoord if you use this option.
It will also cause column names to be printed.
-col#_name='name' if you don't want the default name for the column (which is just col#_name) specifiy a name here.
-form='xxx' instead of -cond. a formula. eg -form='(area/dist+.5)-(area/2)' or -form='myfunc(dist,area)'
not implemented yet.
-line if -cond is satisfied, print the entire line of data (-xcoord and -ycoord not needed, they are ignored).
note: fields will be in the same order than they were in the input files, but formatting may be a bit different.
-xmgr_labels=# add output commands for xmgr which will add a label to each point. # is font size (.5 seems to be a good value).
The command lines all begin with @, so if you have a file produced that way and decide you do not want them
displayed in xmgrace do:
gedmat_to_xy.pl -xmgr_labels=1 gedmat.areas > gedmat.xmgr # you did this thinking you wanted labels
fgrep -v @ gedmat.xmgr |xmgrace -free -pipe # but now you decide you don't want the labels
This command cannot be used with the -col or -line options (ie, -xcoord and -ycoord must be specified).
Unless -unique_header is specified, the label is the patient id number (gotten from the filename field)
and the well id (from the well field) - these are produced by mask_gedmat_images.pl.
If -unique_header is specified the label will be the data line number in the file (excluding blank lines, comments, etc).
health options:
-dummy_health add a dummy field (called dummy_health) for health status (e.g. normal, db1, db2) so I can start testing my stats software.
only use if -col is also specified.
-seed=# seed for random number generator used if -dummy_health is specified. Specifying a seed lets you repeat this over and
over and get the same random numbers. The default is to use the time of day (which will change with each running).
-t2dm=id[:...:id] list of patients which are type 2 diabetic (t2dm). These ids should match the patient id field (the 3 digit number)
in the *.areas files being analzyed. For example in 017_VQU_OM.areas:
filename well type day area farea area_mex area_mex_border dist date id perr aerr bad
B05_017_VQU_OM_Area.zip B05 OM 11 2866176 2869698 2563883 1642634 1712.8 date_sure 017 0 0
So if patient 17 is t2dm specifiy -t2dm=017
-gdm=id[:...:id] list of patients which are gestational diabetic (gdm).
The name should be the (3 digit) id field in the data line, just as in the -t2dm example.
if either (or both) of -tdm and -gdm are specified, a field called "health" will be added to the output. It will
have values t2dm, gdm, or normal.
misc options:
-one_space separate columns by exactly one space so sas can parse it easier (see also: spaces.pl) with proc import.
-silent suppress some messages
-debug stuff to stderr
-Debug other stuff to stderr
-debug_print_columns
examples:
# analyze some zip files
mask_gedmat_images.pl > file.areas
# compare the area mask_gedmat_images.pl produced with that produced by fiji (these should be very close):
gedmat_to_xy.pl -cond='if ($date eq "date_sure") {1;} else {0;}' -xcoord='$area' -ycoord='$farea' file.areas | xmgrace -free pipe
# compare the area mask_gedmat_images.pl produced with area_mex_border (eliminating regions near the well border).
# This is excluding the explant area. ignore any lines with uncertain dates or parse errors (perr) or area errors (aerr):
gedmat_to_xy.pl -cond='if (($date eq "date_sure") && ($perr==0) && ($aerr==0)) {1;} else {0;}'
-xcoord='$area_mex' -ycoord='$area_mex_border' file.areas | xmgrace -free -pipe
# suppose you just have columns of data produced some other way, and no header line, and you just want to do stuff:
gedmat_to_xy.pl -cond='if ($col5>0) {1;} else {0;}' -xcoord='$col5' -ycoord='abs($col5-$col6)'
-unique_header=none yourdatafile | xmgrace -free -pipe
see also:
mask_gedmat_images.pl
/home/lml/krypton/lml/Corvera/Raz
source code in /home/lml/krypton/bin
get_columns.pl
pulls out specified columns from a file. Ignores comment lines. Header for the file must be the first non-comment line.
usage:
get_columns.pl [options] -cols=name:....:name datafile > out.nxy
options:
-linenum precede each line with a linenumber
-noheader do not print out a header line
-noinfo do not print an info line
-cond='xxx' a perl expression to be evaluated to determine which rows of data to pull data from. default is all rows.
if it evaluates to > 0 the row will be used.
e.g., -cond='if ($tissue eq "OM") {1;} else {0;}'
e.g., -cond='if ($date == 7) {1;} else {0;}'
note the use of single (not double) quotes to protect this from the shell.
If this is not specified all data lines will produce an x and y value.
Some simple perl info:
use eq (equal) and ne (not equal) for comparing strings (which are in double quotes).
use == != > >= <= for numeric comparisons. && (logical AND), || (logical OR).
put $ in front of all variables (string variables and numeric variables).
statements are separated by semicolons. Blocks specified with braces.
if ( ) { } else { } where the "else {}" is optional. But don't forget the braces.
-skipvals=#:...:# list of values (or text), of lines which should be skipped if any fields have one of these values
-replacevals=#=#:...#=# if this value (or text) is seen in any field, replace it with the second value
example: -replacevals=unk=-1:20=10 replace unk with -1 and replace 20 with 10
-debug stuff to stderr
example:
get_columns.pl -cols=time:mass:growth datafiles > out.nxy
xmgr -type nxy out.nxy
see also:
graph_xpp2.pl
cull
get_dir_results.pl
This program looks in subdirectories, finds files named something_Results.txt and then reads info from them, These files can be the result of analyzing a projection of the 3D stack (5 z slices) or the result of just analyzing the best slice from the stack. By default only stack projected results files are extracted. Results written to stdout. usage: get_dir_results.pl options: -verbose stuff written to stderr -loc=/some/path/here rather than looking in /storage/big1/raz/GEDMAT, look in this location NOTE: the slice options may be buggy, it is better to use get_dir_slice_results.pl -also_use_slice_data also include Results from single slice data (rather than just stack project data). this may cause multiple entries in the output (one for stack results and one for slice results). -only_use_slice_data exclude any file which are NOT from slice data. examples: get_dir_results.pl > all_results.txt see also: get_dir_slice_results.pl get_find_vesselness_results.pl combine_Raz_and_lml_data.pl combine_with_Raz_curated_data.pl
graph_channel_current.pl
Reformats data from the histograms produced by channel_current for graphing by xmgrace. graph_channel_current.pl [options] *.channel_current | xmgrace -source stdin -linetype=# : 0=no lines, 1=lines[default] -symboltype=# : 0=pts[default] 1=circles -normsum : normalize so sum (area under curve) = 1 -normmax : normalize so the max of the curve = 1 -countmax=# : cap each bin count at #. applied before normsum or normmax. -binmax=# : group all counts in bins > # into that last bin -cum : plot cumulative distribution. applied after all other options. See also: /home/lml/krypton/Zhuge/BKandRyr/analysis5/graphall3.csh
makebeads2rpts.pl
This program takes output from makebeads2 and converts it into an rpts format. A coord from [0,1) is considered in the first pixel; this has a pixel coord 1 since rpts uses 1-indexed values. A coord from [1,2) will have a pixel coord 2, etc. usage: makebeads2rpts.pl makebeads2.out > makebeads2.rpts options: -oldres=# : pixel size used to create makebeads2.out. default = 1nm -newres=# : pixel size for makebeads2.rpts default is the same size as oldres. If newres = oldres, the pixel coords won't change (except for being converted to 1-indexed integers). -project : ignore z value, project all pts onto the first plane -jack=# : instead of just coords of each pt, produce a jack of the specified integer radius (in pixels) -verbose
merge_rpts.pl
Takes output from runall.m, which produces a series of rpts files for a data set, where each rpts file is for a different range of z values, and concatenates the paths together. The result can then be analyzed by paths5.pl. Assumes the files passed are already sorted in z (ie, the first file has paths from z = 1 to 500, the next has paths from z = 501 to 1000, etc.). usage: merge_rpts.pl [options] z1_500_tracks.rpts 2_z501_1000_tracks.rpts z1001_1500_tracks.rpts > all_tracks.rpts options: -dx=# how close (in pixels) the paths must be in x to merge (be considered the same path), default = 0.5 -dy=# how close (in pixels)the paths must be in y to merge (be considered the same path), default = 0.5 -verbose_merging prints merging info to stderr -vverbose_merging very verbose, prints other into about merging. -verbose_paths -verbose_files -silent see also: /home/lml/krypton/Satoshi/runall.m /home/lml/krypton/Satoshi/Results/ note: this could probably be easily modified to skip over short gaps in z by defining a -dz and taking out the test that requires a path to end at the maxz of the file. But it would only skip over gaps exactly at the end of the files, unless the comparisons are extended to also look at other paths in the same file.
modtracks.pl
# takes output rpts from track and modifies it so that format is changed to superimpose on a projection image
# where each time pt is represented by a single (projection) z slice. This lets us view the result for huge time series.
# original format:
# xyzw # time
# new format:
# x y t
# assumes time coords passed are zero indexed.
# usage: modtracks.pl [options] file.rpts > newfile.rpts
options:
-everyplane have each track show up on all z planes (all projections), otherwise
only part of the track will be visible at each time point.
-dup duplicate points so a line from t1 to (t1+1) is completely visible
in t1 and in t1+1 (otherwise half will appear in t1 and half in t1+1).
-color color the tracks (use play with options: -P -1 R -P -2 G -P -3 B -P -4 C -P -5 Y)
-tshift=# add # to the time (z) coord of all pts.
# example (a1.i2i through a3.i2i are each a 4D time series):
# track3dg.linux -a 3 -u .33 .33 .25 5 -h .01 a.vel a1.i2i a2.i2i a3.i2i > a.rpts
# modtrack.pl a.rpts > amod.rpts
# reduceima -sets 10 -P a1.i2i aproj1.i2i
# reduceima -sets 8 -P a2.i2i aproj2.i2i
# reduceima -sets 11 -P a3.i2i aproj3.i2i
# concateima aproj1.i2i aproj2.i2i aproj3.i2i aproj.i2i
# addlines aproj.i2i amod.rpts - |play -P -1 R -P -2 G -P -3 B -P -4 C -P -5 Y -
see also:
trackinfo.pl
objs2bb.pl
#this program takes the object outputfile produced by countobjs and creates a pts file #from it. Each object in the original file gets turned into a wireframe cube of the bounding box. #usage: # objs2bb.pl <file.objs >file_bb.pts #example: # countobjs -k -t 2000 2000 -O cs1_4c_c_objs2.i2i cs1_4c_c.objects2 cs1_4c_c.i2i # objs2bb.pl < cs1_4c_c.objects2 > cs1_4c_c_bb.pts # dave -n 0 -u -kptcolor -K -I cs1_4c_c.i2i cs1_4c_c_bb.pts
path2rpts.pl
This program converts path files as produced by gimp to our standard rpts format. Currently only handles one path, and strips off any identifying name. path2rpts.pl [options] infile.path > outfile.rpts options: -xmgr: output xmgr format instead of rpts format
pgfocus2.pl
takes all the data files of readings from Karl's pgfocus calibration routine (e.g., /storage/big1/kdb/pgFocua/20130912/cal_4.txt) and combines them based on calibration voltage, sends them to xmgrace, all the linear array curves for the same voltage on the same graph. pgfocus2.pl options: -files=file1.txt:file2.txt one or more files to analyze, separated by colons. default = all *.txt files in the current directory. -nocoms don't print a curve of the center of masses -keepsets when moving to the next screen of data (next calibration voltage) keep old sets around. -print_stats prints mean center of mass and sd center of mass to stderr as each xmgr page is displayed. -debug text goes to stdout instead of xmgrace see also: pg_focus.pl this puts all the linear array curves for a given run (file) (ie, at many voltages) on one graph. call_pgfocus.pl this calls pg_focus.pl for each file in a directory.
plotdata.pl
Takes sas output from running proc tpsline (see runtest.sas) and graph it. plotdata.pl output.txt > output.xy options: -alldata : tspline putting out all the variables it can (so there are more cols in output.txt that must be parsed) -numbins=# : number of bins for histogram of residuals. this goes to stderr. default = 20 -range=#:# : range for the histogram of residuals. this goes to stderr. default = -10:10 -use_coef : to check my interpretation of what the fitting coefficients mean. This should produce the fitted spline function. See p. 7074 SAS_STAT_Users_Guide.pdf (use okular) and the README entry for 1/25/11. Doesn't seem to be correct at all ??? -print_diag : print to stderr diagonal values of A (a_ii). This is used to calculate the width of the error bounds. must use -alldata with this. xmgrace output.xy In xmgr set the first graph to have no lines connecting pts and a circle symbol (it is a scatter plot).
read_hist.pl
read output from pt_distances, convert histogram to xy column format.
read_hist.pl [options] > histo.xy
options:
-z=minz:maxz:delta only print histograms for these z values
-im1 print histogram for image1, default = all histograms (im1, im2, im1vs2, im2vs1)
-im2 print histogram for image2
-im1and2
-im2and1
-norm=# normalize histogram to sum to #
-nummax=# print nummax to stderr, divide nummax by # before printing (e.g., # could be area of cell to yield rate/area)
this will also compare the observed mean minimum separation with that predicted from random placement
(only correct comparison if # used is the area). This info also added as comments to the stdout.
Might be off by a bit since the .9 bin is really a distance of .9 to 1, ie, really 1 pixel away.
-spacing=# spacing between histogram entries, default = .1 pixels
-avg=# rather than printing straight histogram, do a running average of the # preceeding histograms (first #-1 histograms
produced won't be an average of # histograms). running average is of all z values in the specified set, even
if only a subset is printed (and you must specify exactly one of -im1, -im2, -im1and2 -im2and1 to use this).
This option also causes statistics (like avg minimum distance) to be based on the running average histogram.
-nozero
-debug
example:
# print histogram for im1 vs im2 for z value 1 to 500 by 100
max2d_simple -p glut.rpts glut.i2i
max2d_simple -p clathrin.rpts clathrin.i2i
pt_distances -h glut.rpts clathrin.rpts > glut_vs_clathrin.dat
read_hist.pl -im1and2 -z=1:500:100 < glut_vs_clathrin.dat | xmgrace -source stdin
(for some reason my xmgrace isn't taking pipes from read_hist.pl, so write to file instead:
read_hist.pl -im1and2 -z=1:500:100 < glut_vs_clathrin.dat > tmp.xy; xmgrace tmp.xy)
ReadROI.pl
This program reads an *.roi file from stdin, as produced by ImageJ or fiji (Analyze->Tools->Roi Manager -> More -> Save As ...)
and prints the x y coordinate pairs to stdout. This could be piped into xmgr (/usr/bin/xmgrace -free -pipe) for visualization.
I may eventually use this to go back into the image file at these locations, extract new features at those locations
and create my own *.arff files for the Plugins->Segmentation->Advanced Weka Segmentation to use for training and
then classification.
Note: roi files with more than about 250,000 x y coordinate pairs will be too big and might get truncated in funny ways.
usage:
ReadROI.pl [options] < file.roi | xmgrs
options:
-addone convert from 0 indexed to 1 indexed
-rpts=objname:# write out in rpts format, sets -addone option. # is the w value (typically -1).
-readcoords=# force a read of this number of coordinate pairs (e.g., (x,y) pairs).
see -verbose output -> "max possible coords" for the largest number you should specify.
-verbose prints header info to stderr
-debug some info prints to stderr
-diag print all the x coords, then all the y coords (and in relative coords), this should
match up with: od -x --width=2 -A d --output-duplicates --skip-bytes=64
but you will need to use bc with ibase=16 and then reverse pairs of digits (and make
letters caps) to convert the od values to the -diag values.
-relative rather than print the absolute coords, print relative to the starting position (?)
source code in:
~/krypton/Corvera/grant2010
See also:
~/krypton/Corvera/grant2010/README for 7/24/12
~/krypton/packages/fiji/src/CallRoiDecoder.java
~/krypton/bin/Jrun_CallRoiDecoder
~/krypton/bin/mask_gedmat_images.pl
rgbtif2i2i.pl
note: this version of converttif2.pl handles only rgb tif files. This is different than some of the other directories. in this directory: rgbtif2i2i.pl -threedigits [options] will convert and concatinate files named root.TIF to a single file called root3dtag.i2i where "root" is as defined by -root option and "tag" is defined by -tag option options: -root=rootname : converts files numbered rootname#.TIF, rootname##.TIF and rootname###.TIF, default rootname is human_explant_p33_s1_001_w1brightfield_t note: a space in the name will be replaced with an underscore (_) on output. -tag=tagstring : default is the empty string; -threedigits : all numbers in the name have 3 digits, e.g., root001.TIF rather than root1.TIF -twodigits : all numbers in the name have 2 digits, e.g., root01.TIF rather than root1.TIF -dims=xdim:ydim : dimension of images. default = 640:480 -tif : use .tif instead of .TIF as the extension. -start=# : number to start at, default = 1 -stop=# : number to stop at (inclusive), default = 999. It will stop before this if no more valid files are found. note: if some of the files in the range start to stop are missing, the conversion program will continue until the stop # specified is reached (but if -stop=# not specified then the conversion will stop). -channel=red|blue|green which channel to pull out. default = red -verbose : print what does as it goes (if -debug, it will still print, but won't do anything) -chunk=# : number of files to concatenate together at at time. default = 20 -debug : don't do anything. combine with -verbose to see what it WOULD do. example: convert_rgbtif.pl -tif -threedigits -start=2 -stop=576 -root='48_hr_human_explant001_3 '
rpts2i2i.pl
Convert rpts file into an i2i image. usage: rpts2i2i.pl [options] file.rpts file.i2i options: -w=# intensity the pixels should be set to. default is that specified file.rpts, or -1 if not specified there. -bkgnd=# value of background pixels. default = 0. -image=image.i2i create an image the same size as image.i2i. default = 100, 100 , 10 -dims=#:#:# set the image size to xdim:ydim:zdim -verbose info to stderr See also: morphrpts_newpaths, addlines, histogram2, maskedsmul
rpts2jack.pl
Convert rpts to jack.rpts usage: rpts2jack.pl -jack=# <in.rpts > out.rpts options: -twod make a 2d jack not a 3d jack -jack=# # is the half-width of the jack (default = 1). -w=# # is the w value printed out, default = -1 (or the incoming value if it exists in the file). -break_on_newline addlines seems to like this -verbose
rpts2rpts.pl
rpts2rpts.pl converts one rpts file into another. usage: rpts2rpts.pl file.rpts > newfile.rpts options: -dx=# : amount to add in x, default = 1 -dy=# : amount to add in y, default = 1 -verbose
rpts2xmgr.pl
Takes an input rpts file (as produced by planimeter) and writes out the specified objects into an xmgr file format. rpts2xmgr.pl -name1=#-# [-name2=#-# ...] < in.rpts > out.xmgr or rpts2xmgr.pl -name1=#-# [-name2=#-# ...] in1.rpts [in2.rpts ...] > out.xmgr example: rpts2xmgr.pl -foo=1-5 -bar=2-8 < in.rpts > out.xmgr This will take the zslices associated with the object named foo (slices 1 through 5) and convert them into xmgr format (z dimension will be lost). Similarly for the object named bar. note: this is untested and unlikely to be able to handle any but the simplest files.
stack2rpts.pl
Reads in file produced from DAVE crosshair stack, converts it to rpts format. Reads stdin, writes stdout. May be off by one (zero indexed instead of 1 indexed). options: -color: color the paths to preserve parent/child relationship (colors: 0-6) -jack=#: print a jack at each place a cell splits. # is the radius of the jack.
text2i2i.pl
This will take data produced by Karl's True Focus Ardueno and convert into an image for diagnostic purposes.
Reads in a text file. Each line of the text file has (normally) 128 pixel values and a last value (usually the 129th)
which is a center of mass coordinate to go to an rpts file. Produces an image and an rpts file.
Use addlines2 to combine outfile.rpts with outfile.i2i (it handles floating point values).
usage:
text2i2i.pl [options] infile.txt outfile.i2i outfile.rpts
options:
-round produce integer output by rounding.
-add=# add # to the com coordinate. Some of Karl's earlier software produced a com number 2 too large, so
-add=-2 would be used to correct for that.
-debug
example:
text2i2i.pl infile.txt outfile.i2i outfile.rpts
addlines2 outfile.i2i outfile.rpts - |xmgrace -free -pipe
cp outfile.rpts tmp.xy
vi tmp.xy # get rid of text at the top. delete the last column (:.,$s/-1//)
cull -2 -1 < focus5.tmp |xmgrs # then doing a 30 pt running avg, then 15 drop pts from the start and the
# end of the original set so they are the same size. Then select the first set and do:
# y = y - S1.y to see the "noise" in the system. 1 pixel represents 100 nm focus shift (I think).
see also:
chres_rpts.pl, chres # if you want to see the rpts in higher resolution. At the original resolution you sometimes see "wiggles"
# in the addlines path which aren't real but just due to the integer nature of pixels.
vals2xmgr.pl
this program takes the output of printvals (when used to create one xline per zslice through the image as shown below), calculates changes from z=0 (ie, time zero) unless -abs is specified, and reformats the output as a series of curves for xmgr (can also be read into matlab via readxmgr). This is intended to be used to get concentration as a function of distance from a channel (located at (0,#1)) from images produced by cellsim. printvalsL -y #1 #1 image.i2i > file.vals (where both numbers passed to the y option are the same) vals2xmgr [options] < file.vals > file.xmgr options: -pixsize=#: pixel size, default = 0.1 -abs: just reformat, keeping absolute values
xmgr2rpts.pl
Converts xmgr formatted files to rpts format. Only handles very simple ones. Meant to convert from output from gvf snake to rpts format. Converts floats to ints. Note: you need to delete any header lines from the top of the xmgr file. It can also only have one data set in it. xmgr2rpts.pl < file.xmgr > file.rpts options: -round: round instead of truncate.
con2rpts
Convert a gsnake contour (.con) to an rpts format
Usage: con2rpts conIn.con conOut.rpts -I image.i2i
-I image.i2i : specifies the image file on which the outline is superimposed
createBoundary
createBoundary [options] image.i2i data.rpts boundary.i2i
Options:
-o : points in rpts file marked outside image (default inside)
Example:
createBoundary image.i2i data.rpts boundary.i2i
image.i2i : image from which rpts was drawn
(long axis of cell in z direction)
data.rpts : Rpts data file (drawn from input image)
boundary.i2i : output Boundary image ready for histogram
Keywords:
intensity histogram, floodfill, boundary, distances.
Other programs:
boundary, flood, planimeter, histogram, transp
\n
cyl2cart
expand_projection
expand_projection takes a projection image produced by reduceima with the (undocumented)
-when option and turns it into a 3D image, with pixels at the z values specfied by the whenimage.i2i
set to 1 and the others to 0.
Usage: expand_projection whenimage.i2i [image.i2i] image3D.i2i
options:
image.i2i use the intensity values from image.i2i (usually the original image), rather than setting
the values to 1
-s #: image.i2i has been segmented so its first slice is really slice # (0-indexed).
e.g., if whenimage.i2i has a pixel value of V, pixel at z = V-# in image.i2i will be used.
-v #: rather than setting the "when" pixels to 1, set them to #
-z #: set the zdim of the new image to #, rather than the max pixel value in the "when" image.
-d debug
example:
find which z plane the toxin (green_tb) maxima occurs, only look at a subset of the 1800 z slices:
reduceima -when -Z 500 1500 1001 -H 101208_4_green_tb_ng.i2i 101208_4_green_tb_ngPwhen.i2i
expand_projection -z 1800 101208_4_green_tb_ngPwhen.i2i 101208_4_green_tb_ng_max3D.i2i
does the toxim show up in the cell from the Tf (red_tf) sites?
dave -I 101208_4_red_tf_ng.i2i -I 101208_4_green_tb_ng_max3D.i2i
example:
segment -Z 500 1500 1 101208_4_green_tb_ng.i2i 101208_4_green_tb_ng_z500_1500.i2i
expand_projection -z 1800 -s 500 101208_4_green_tb_ngPwhen.i2i 101208_4_green_tb_ng_z500_1500.i2i 101208_4_green_tb_ng_max3D.i2i
does the toxim show up in the cell from the Tf (red_tf) sites?
dave -I 101208_4_red_tf_ng.i2i -I 101208_4_green_tb_ng_max3D.i2i
see also: ~/krypton/Zhuge/toxin/README 8/10/12
source code in: /home/lml/krypton/facil
im2pts
im2pts takes images produced by Kevin and converts them into ascii lists of points
suitable for reading into the matlab tracking program.
image1 is 0 except where an object has been found, there it has an object id.
Kevin tracks the object, so the same object in adjacent time pts (z slices) will
have the same id (if correctly tracked).
image2 (if specified) is the original data, so that information about the object can be
extracted. This extra info should help the matlab track program perform better.
Usage: im2pts image1 [image2] outfile
options:
-o: want output info to be 1-indexed (default = 0-indexed).
-i newimage radius: produce a newimage by keeping a region of radius around each id pixel.
image2 must be specified too. For visualization after track_vesicles.m
-I inner outer: when image2 specified, calculate the iod within +/- inner pixels after subtracting
the average background from each value. The background is the average value in the
pixels in the outer region (i.e., +/- outer pixels, excluding inner pixels).
If not specified, the pixel value from image2 at just the point location is used.
-v #: level of verbosity, default = 0. larger integers produce more output.
see also: ~/krypton/tracking/ModifiedMHT/tracking/test_tracking_script.m
/home/lml/krypton/Corvera/paper2010/track_vesicles.m
source code in: /home/lml/krypton/Corvera/paper2010
init_cyl
Incorrect command line. Usage: init_cyl [options] arg1.rpts ... arg2.i2i Files: arg1 rpts file ... additional rpts files arg2 output image file options: -c: create a sphere (default cylinder) takes one or more rpts files and puts them in the proper format for min_cylinder. note: image size is taken from the first rpts file additional rpts files overwrite data from previous rpts files in output image. note: entire image name (with extension) is required. note: a - in place of an image name means stdin or stdout.
make_surface
Usage:
make_surface [options] mesh.i2i out.rpts
Description:
This program takes an input mesh file (as produced by
shrink_wrap, min_surface or min_plane) and converts it to a
surface, then writes it out as a points file in a format that
is compatable with addlines.
Expects: A mesh file from shrink_wrap.
Modifies: Nothing.
Returns: A points file of all the points on the surface
described by the mesh file.
Options:
-I name.i2i: also produce a 3D .i2i image with surface voxels set nonzero.
see options -c and -D below for modifications of this image.
-c # : color (intensity) of surface voxels of output image file
(use with -I option). default = -2 at wireframe locations
and -1 at other surface voxels. If -c is specified all surface
voxels get set to that color (wireframe and other surface voxels).
-O name.pts: surface vertices output as rpts file.
-Z : only draw lines between pts in the same z slice.
this is used for time series data.
-P : surface mesh is for a plane, not a cylinder
so do not connect the last point to the first.
-D # # # : max xdim, ydim and zdim of output.
If you want your output image (-I option) to be
a particular size you should set it here, otherwise
the size is the maximum dimensions of the surface.
-i : history line for output image.
-v : verbose, prints to stderr how many triangular patches were
seen (and any degenerate patches which were lines or pts).
-h : print this message.
Caveats:
There may be a few very small holes in the surface.
Notes:
Entire image name (with extension) is required.
A - in place of an image name means stdin or stdout.
If a compressed or packed image is given, this program
will read it, given that there is sufficient space in /usr/tmp.
Keywords:
surface shrinkwrap shrink_wrap addlines boundary
Additional programs:
shrink_wrap dave addlines
merge_rpts
Usage: merge_rpts [options] arg1 arg2 ... Takes several rpts files merges them into one. If a z= seen Only points in last zslice kept. options: note: entire image name (with extension) is required. note: a - in place of an image name means stdin or stdout.
mesh2rpts
Usage: mesh2rpts [options] meshfile rptsfile Description: Takes a mesh file created by shrink_wrap and converts it to a rpts file compatible with planimeter. Options: -o name: name of object in the mesh file. Keywords: shrink_wrap meshfile rptsfile dave Notes: Entire image name (with extension) is required. A - in place of an image name means stdin or stdout.
morphrpts_newpaths
0, 1 Incorrect number of arguments. Usage: morphrpts_newpaths original.rpts output.rpts Description: Erode or dilate an outline in a points file Options: -s xdim ydim zdim: default 100 100 10 -S size : amount to move pts. Default 6 -e : erode points (make outline smaller) -d : dilate points (make outline larger) -h : display this message -v : verbose Keywords: Programs: morph maskedsmul addlines image2pts create_dcim
pts2i2i
pts2i2i takes a list of pts as produced by stats1.pl and turns them into an image pts2i2i: inpts outimage.i2i options: -x xdim: outimage should have the specified xdim. Otherwise it will be just big enough to include all the pts in the inpts file. -y ydim: same for ydim -z zdim: same for zdim -r #: instead of a single pixel, put down a square of the specified radius. 1 means +/-1, i.e., a 3x3 region. 2 means 5x5, etc. examples: count_2d_objs.pl -allstats=file.allstats file_mask.i2i file_mask.count_2d_objs stats1.pl file.allstats > file.allstats2 tracking/ModifiedMHT/tracking/test_tracking_script.m iminfo -d file_mask.i2i # to get xdim ydim zdim pts2i2i -x xdim -y ydim -z zdim file.allstats2 file_spots.i2i see also: ~/krypton/Corvera/paper2010/Dynamics_in_Dave/README source code in: /home/lml/krypton/facil
Graphing
avg_curve2.pl
avg_curve2.pl similar to avg_curve.pl, but combines all the input data files into one curve with sem error bars (instead of producing one curve per input file). reads output of coloc_tseries2 for a bunch of files and calculates an average curve with error bars. options: -debug example: # runall.pl has already created the *.txt files analyzed below avg_curve2.pl *before*_coloc2_r1.txt > coloc2_r1.xydy xmgr -settype xydy coloc2_r1.xydy
boxandwhisker.pl
parse data line like (one header line, the rest data lines):
patient tissue well explant rate day dummy_health
003 OM B02 595424 79067.2857142857 7 db1
prints out rate data in a format suitable for xmgr's settype XYBOXPLOT (X, median, upper/lower box, upper/lower whisker)
Where X is the patient id number.
usage:
boxandwhisker.pl [options] [filename| <filename]
options:
-cond='xxx' a perl expression to be evaluated to determine which rows of data to analyze. default is all rows.
if it evaluates to > 0 the row will be used.
e.g., -cond='if ($tissue eq "OM") {1;} else {0;}'
e.g., -cond='if ($date == 7) {1;} else {0;}'
note the use of single (not double) quotes to protect this from the shell.
If this is not specified all data lines will produce an x and y value.
Some simple perl info:
use eq (equal) and ne (not equal) for comparing strings (which are in double quotes).
use == != > >= <= for numeric comparisons. && (logical AND), || (logical OR).
put $ in front of all variables (string variables and numeric variables).
statements are separated by semicolons. Blocks specified with braces.
if ( ) { } else { } where the "else {}" is optional. But don't forget the braces.
-offset=# add # to the X position of each data pt
-quant=#:#:#:#:# default quantiles are 0:.25:.5:.75:1 (.5 = median, 0 and 1 are lower and upper whisker, .25 and .75 lower/upper box)
(I am not sure how perl interpolates if the exact quantile is unavailable or duplicates are present)
-debug stuff to stderr
example:
boxandwhisker.pl gedmat_dummyhealthr.data |xmgrace -settype xyboxplot -free -pipe
# look at OM and SQ data split out. Offset one relative to the other for easier viewing:
boxandwhisker.pl -cond='if ($tissue eq "OM") {1;} else {0;}' gedmat_dummyhealthr.data > tmp.OM
boxandwhisker.pl -cond='if ($tissue eq "SQ2") {1;} else {0;}' -offset=.3 gedmat_dummyhealthr.data > tmp.SQ
xmgrace -settype xyboxplot tmp.OM tmp.SQ
# look at all the data, but try to exclude the very extremes. compare it with the original
boxandwhisker.pl gedmat_dummyhealthr.data |xmgrace -settype xyboxplot -free -pipe
boxandwhisker.pl -quant=.05:.25:.5:.75:.95 -offset=.3 gedmat_dummyhealthr.data > tmp.box
# in xmgrace: Data->Import->Ascii, Set type: XYBOXPLOT, and import tmp.box. set
# Plot->Set Appearance turn "line properties" to None for each set.
see also:
boxandwhisker2.pl same as this script, but a bit more flexible.
xmgrace --help
http://plasma-gate.weizmann.ac.il/Grace/doc/UsersGuide.html
/home/lml/krypton/Corvera/Raz/gedmat_dummyhealthr.data
/home/lml/krypton/Corvera/Raz/README
gedmat_to_xy.pl used to produce gedmat_dummyhealthr.data. can also pull out arbitrary rows and columns
coloc.pl
calls coloc3way with a bunch of different thresholds. Creates an output file which can be
graphed with VennDiagram.m.
coloc.pl im1.i2i im2.i2i im3.i2i > out.coloc3way
options:
-time_vs_thresh just generate a file which shows which times correspond to which thresholds. images can be dummy names.
-t1=#:#:# threshold one should go from # to # by #. default = 800:1200:50
-t2=#:#:# threshold two should go from # to # by #. default = 800:1200:50
-t3=#:#:# threshold three should go from # to # by #. default = 2000:4000:200
-numthresh=#1:#2:#3 ignore the third number in the -t1, -t2, and -t3 options (or in the default) and instead calculate a
threshold step size so that #1 thresholds are applied to im1.i2i (dat image)
#2 thresholds are applied to im2.i2i (ctx image), #3 thresholds are applied to im3.i2i (rin image).
e.g., -numthresh=5:10:10 will apply 5 different thresholds to im1.i2i, and 10 thresholds to im2.i2i and im3.i2i.
Using this option lets me insure that each call to coloc.pl will produce exactly the same number of data points;
this may make the statistical comparison between cells easier.
-Melikian=file in addition to creating output for VennDiagram, create text file with the coloc percentages
Dr. Melikian is interested in. Graph via xmgr -nxy. See header in file for coloc info.
For this to work the three image files MUST be specified in the order 405 594 488 (i.e., rin.i2i ctx.i2i dat.i2i)
-Melikian2=file Same as -Melikian, but dat (488) is image 1, ctx (594) image 2, and rin (405) image 3
-area=file.xyr also print out Melikian data as xyr where r is proportional to the number of pixels involved
(i.e., the denominator used when calculating the percent colocalized).
prints to "file.xyr". Must also specify either -Melikian or -Melikian2
xmgrace -graphtype fixed -settype xyr file.xyr
-matlab=file:# like -area but as columns so easy to read into matlab: t p1 n1 p2 n2 p3 n3 p4 n4
# is the max n value (ie, all n values are scaled so the max over n1,n2,n3,n4 is #).
Since p1,p2,p3,p4 are percent, # is typically set to a small part of that, e.g. .05.
If # is negative then abs(#) is just used as a scale factor (so -1 would just keep the
n values as the number of voxels). Graph in matlab with line width of 2*n1:
matlab # import data=(tdim,9)
>> lowy = data(:,2) - data(:,3); hiy = data(:,2) + data(:,3); x = data(:,1);
>> area(x,hiy,'FaceColor',[1 0 0],'EdgeColor','r','LineWidth',1); hold on;
>> area(x,lowy,'FaceColor',[1 1 1],'EdgeColor','r','LineWidth',1);
Must also specify -Melikian or -Melikian2.
-debug
-verbose
example:
coloc.pl im1.i2i im2.i2i im3.i2i > coloc.data
coloc.pl -time_vs_thresh im1.i2i im2.i2i im3.i2i > coloc.times
matlab
>> VennDiagram('coloc.data','percent') # and manually look at coloc.times while doing this to convert times to thresholds.
see also:
~/krypton/packages/fiji/Fiji.app/plugins/jacob_.jar - more traditional colocalization measures
coloc_tseries2
colocalizeL
some_stats.pl
parse data line like (one header line, the rest data lines):
patient tissue well explant rate day dummy_health
003 OM B02 595424 79067.2857142857 7 db1
prints out rate data in a format suitable for xmgr's settype XYBOXPLOT (X, median, upper/lower box, upper/lower whisker)
Where X is the patient id number.
usage:
boxandwhisker.pl [options] [filename| <filename]
options:
-cond='xxx' a perl expression to be evaluated to determine which rows of data to analyze. default is all rows.
if it evaluates to > 0 the row will be used.
e.g., -cond='if ($tissue eq "OM") {1;} else {0;}'
e.g., -cond='if ($date == 7) {1;} else {0;}'
note the use of single (not double) quotes to protect this from the shell.
If this is not specified all data lines will produce an x and y value.
Some simple perl info:
use eq (equal) and ne (not equal) for comparing strings (which are in double quotes).
use == != > >= <= for numeric comparisons. && (logical AND), || (logical OR).
put $ in front of all variables (string variables and numeric variables).
statements are separated by semicolons. Blocks specified with braces.
if ( ) { } else { } where the "else {}" is optional. But don't forget the braces.
-offset=# add # to the X position of each data pt
-quant=#:#:#:#:# default quantiles are 0:.25:.5:.75:1 (.5 = median, 0 and 1 are lower and upper whisker, .25 and .75 lower/upper box)
(I am not sure how perl interpolates if the exact quantile is unavailable or duplicates are present)
-debug stuff to stderr
example:
boxandwhisker.pl gedmat_dummyhealthr.data |xmgrace -settype xyboxplot -free -pipe
# look at OM and SQ data split out. Offset one relative to the other for easier viewing:
boxandwhisker.pl -cond='if ($tissue eq "OM") {1;} else {0;}' gedmat_dummyhealthr.data > tmp.OM
boxandwhisker.pl -cond='if ($tissue eq "SQ2") {1;} else {0;}' -offset=.3 gedmat_dummyhealthr.data > tmp.SQ
xmgrace -settype xyboxplot tmp.OM tmp.SQ
# look at all the data, but try to exclude the very extremes. compare it with the original
boxandwhisker.pl gedmat_dummyhealthr.data |xmgrace -settype xyboxplot -free -pipe
boxandwhisker.pl -quant=.05:.25:.5:.75:.95 -offset=.3 gedmat_dummyhealthr.data > tmp.box
# in xmgrace: Data->Import->Ascii, Set type: XYBOXPLOT, and import tmp.box. set
# Plot->Set Appearance turn "line properties" to None for each set.
see also:
xmgrace --help
http://plasma-gate.weizmann.ac.il/Grace/doc/UsersGuide.html
/home/lml/krypton/Corvera/Raz/gedmat_dummyhealthr.data
/home/lml/krypton/Corvera/Raz/README
gedmat_to_xy.pl used to produce gedmat_dummyhealthr.data. can also pull out arbitrary rows and columns
More Utilities
gimp2map.pl
create a colormap for the Linux version of DAVE from a GIMP color palette
first create a GIMP color palette:
get into GIMP (K->Graphics->GIMP)
create a new image (File->New)
pop up the gradient editor (Right click in image window->Dialogs->Gradients...)
create a new gradient (Edit, then New Gradient)
convert the gradient to a palette
(Right click in image window->Dialogs->Palette...
Import
Type in the name of the gradient you created above, sample size should be 256
Import (type gradient, which is the default)
Save - this will put it in .gimp-1.2/palettes directory)
then:
gimp2map.pl [options] < gimp_pallete > mymap.map
options:
-map=# map number. default = 15;
see also:
makemap.pl: an alternative way to create the maps, but you don't see the colors as you do it.
K->Graphics->KColorEdit, K->Graphics->KColorChooser : but these don't automatically interpolate
between chosen colors, but you could edit the files (with Kate) and then apply makemap.pl
makemap.pl
create a colormap from linear color pieces. makemap.pl [options] < mapfile > mymap.map options: -map=# map number. default = 15; A mapfile looks something like this (you should define it from 0 to 255): #this is a mapfile 0 white 1 black 60 0 0 255 120 cyan 180 green 200 yellow 220 yellow 255 red Either the second column is a color name (white,black,red,green,blue,yellow,cyan, or purple) or it is an rgb triple, with each color having a value from 0 to 255. The resulting mymap.map can be read into dave via the -loadmap option. see also: color.pl
paste.pl
Paste together columns of data from different files. Each file must consist of a fixed number of columns (i.e., each row in the file has the same number of columns). Each file can have a different number of columns and a different number of rows. If a file runs out of rows (i.e., has fewer rows than the file with the maximum number of row) its fields will be filled with a field variable, so the number of columns is preserved. Output is written to stdout. usage: paste.pl file1 file2 ... filen > files options: -fv=# : field variable to use. # can be any string. default = -1 -verbose : info to stderr
addconstant
This program adds a constant to an image. Usage: addconstant [options] inputimage outputimage options: -a #: add the specified value. default = 0 -m #1 #2: min value allowed. anything below #1 will be set to #2. default = -32767 and -32767 -M #1 #2: max value allowed. anything above #1 will be set to #2. default = 32768 and 32768 -v : verbose, stats on how many pixels below the min or above the max written to stderr see also: rescale note: compressed images are temporarily uncompressed into /usr/tmp.
bsubtract
bsubtract does background subtraction using 2 background images. inimage.i2i is must be composed of 2 alternating groups of images (e.g., different wavelengths). back1.i2i is a 2D image subtracted from all group1 images, back2.i2i from group2 images. Handles short ints (our normal format) and floats. Usage: bsubtract [options] inimage1.i2i back1.i2i back2.i2i outimage.i2i Options: -v: verbose -n: allow results to go negative -g # #: number of 2D images in group1 and group2, default = 120 and 40 Bugs: The created image has a strange permission. fix it by doing: chmod 644 outimage.i2i User must use imsets to set tdim correctly afterwards. Source code in ~krypton/lml/facil
colocalizeL
Create a colocalized image from two input images. Colocalization determined on a pixel by pixel basis.
Usage: colocalize [options] im1.i2i im2.i2i coloc.i2i
options:
-t # #: thresholds. voxels colocalize if im1 > #1 and im2 > #2 (0 and 0 defaults).
-b: binary output. colocalized voxels set to 1, others to 0
(default sets colocalized voxels to max of the two.
-v #: set colocalized voxels to # (default is max), others to 0.
note: entire image name (with extension) is NOT required.
note: a - in place of an image name means stdin or stdout.
note: a compressed image will be automatically be uncompressed.
colorize_image
Takes an image and colorizes it. produces red, green and blue images. Display them in dave with all 3 channels the
same scale factor.
Reads and writes a z slice at a time so should work on huge images.
Usage:
colorize_image [options] inimage.i2i redimage.i2i greenimage.i2i blueimage.i2i
Options:
-c colorindex.i2i: use the values in colorindex.i2i (modulo 7) as one of 7 colors
if -c not specified, values in inimage.i2i are used both for intensity and color info.
-s scale black: inimage.i2i -> newval = (oldval-black)/scale
. default = autoscale to max of 255 -v: verbose
�xamples:
objs_via_ascent -t 100 in.i2i in_id.i2i > in.count
colorize_image -c in_id.i2i in.i2i inr.i2i ing.i2i inb.i2i (autoscales so max will be 255)
dave -S all 1 0 -nocomp -I ing.i2i -I inr.i2i -Iinb.i2i (then turn blue channel on in dave)
Source code in ~krypton/lml/facil
Compute
Usage: Compute [options] arg1 arg2 arg3 options: -a: add two images. -s: subtract two images. -m: multiply two images. -d: divide two images. -h: prints this message. note: Args require entire image name (with extension). note: A - in place of an image name means stdin or stdout. note: Images have to be of the same dimensions. note: Images can be of type real or short int, the note: resulting image will be promoted appropriately.
create_dcimL
Creates a dc image. Usefull since addnoise does not let pixel values go < 0 which skews the statistics. So start with a dc offset. Usage: create_dcim [options] outim.i2i options: -d dc_val: dc value. default = 100 -s xdim ydim zdim: image size, default = (100,100,1) note: entire image name (with extension) is required. note: a - in place of an image name means stdin or stdout. note: see also makedcim.
extract_vals
extract_vals takes output as produced by find_rpts_path.pl, e.g. (x,y,z,w) one per line, and pulls the voxel values for those coords from the specified image file. note: you may have to delete header lines from coords.txt. Usage: extract_vals image.i2i coords.txt > vals.txt options: -o: coords.txt is 1-indexed. I think this is what find_rpts_path.pl produces. -v: verbose. additional output to stderr. see also: /home/lml/krypton/Corvera/paper2010/fig1/README 7/1/10 source code in: /home/lml/krypton/facil
extract_huge
Lets you extract part of a huge image (>2GB, hopefully). It only reads in a z slice at a time, so doesn't use much
memory. Reads and writes sequentially without closing a file, so hopefully should work on pipes too.
Note: right now the largest image that can be read in can have 100000 2d images altogether
Usage:
extract_huge [options] inimage.i2i outimage.i2i
Options:
-p #read #skip #read ...: read # zslices, then skip # zslices, then read # z slices, etc.
this pattern will be used over an over to read z slices from inimage.i2i
-z startz stopz: these are the zslices which get read in from inimage.i2i. 1-indexed, inclusive.
if stopz = -1 that means go to the end(-z 0 -1 is the default)
-r: replace the skipped zslice with the last previously read zslice
-v: verbose (place before -p on command line to see -p parsing)
example:
starting at the 10th z slice until the end, read (and keep) 5 zplanes, then skip 3, then keep 2, skip 3 ...
extract_huge -z 10 -1 -p 5 3 2 3 in.i2i out.i2i
Source code in ~krypton/lml/facil
fit60hz
This program fits 60hz (noise) to input data sampled at a given rate. It prints out the 60hz waveform and the data minus the 60hz waveform (xmgr format) to outputfile. Input should be pairs of floats (time_in_seconds amplitude), other columns (and # lines) ignored. If outputfile is the same as inputfile it will append to the end of it. Usage: fit60hz [options] inputfile outputfile options: -m : first column is in mseconds, not seconds (output will be in msec too). -s #: sampling rate (samples/sec). If this is specified then it is assumed that the first column of data is just a counting variable, not time in seconds. -d # #: only use data with column 1 values between these numbers (inclusive) for the fit. -f: data is in 5 columns, column 1 is time (use -s in addition if it is count) and column 3 is the data. Lines whose first character is # are ignored. -p amp phase dc: don't fit, just subtract this (y = Amp*sin(60t+phase)+dc) from the data. phase in degrees. -D dc: fix the dc value at the specified value. [source code in /usr/people/lml/voxel/JSinger/60Hz directory]
float2intL
This program converts a floating point image format into a short int (ie, "normal") image format. Usage: float2int [options] inimage.i2i outimage.i2i options: -S #: just muliply all values by #. -m #: rescale so new min = # (default = 0) -M #: rescale so new max = # (default = 10000) -s: keep the original scale (ie, same min & max as the original image). -q: quiet mode. don't print message for each voxel scaled out of range. -v: print extra info, verbose. note: entire image name is NOT required. note: a - in place of an image name means stdin or stdout.
iminfo
Reads a short or float image and prints to stdout (coords are 0 indexed): xdim ydim zdim minval minx miny minz maxval maxx maxy maxz type where type is either INT (for short int) or REAL (for 4 byte floating point) Usage: iminfo image.i2i options: -d: only print xdim ydim zdim, not all the other info. -x: only print xdim, not all the other info. -y: only print ydim, not all the other info. -z: only print zdim, not all the other info. note: entire image name (with extension) is NOT required. note: a - in place of an image name means stdin or stdout. note: a compressed image will be automatically be uncompressed.
mask_image
Erases all voxels not under a mask image voxel. Works on char, short int, and float images. inimage.i2i and maskimage.i2i must be the same x and y dimensions. If maskimage.i2i is not the same zdim, then just the first z plane of maskimage.i2i is applied to all the z planes of inimage.i2i (i.e., maskimage.i2i is used as a 2D mask applied repetitively). All images must be the same format. Usage: mask_image [options] inimage.i2i maskimage.i2i outimage.i2i Options: -r newval: replace voxel in inimage which is under the mask with newval (default = original value) -o newval: replace voxel in inimage which is NOT under the mask with newval (default = 0) -m maskval: any voxel > maskval in maskimage.i2i is considered a mask voxel (default = 0) -M maskval: any voxel = maskval in maskimage.i2i is considered a mask voxel. -b minval maxval: any voxel > minval and < maxval in maskimage.i2i is considered a mask voxel (default = 0) -B minval maxval: any voxel >= minval and <= maxval in maskimage.i2i is considered a mask voxel (default = 0) source code in /home/lml/krypton/facil see also: mathi2i, maskedsmul
max2d
This program finds voxels which are > all 8 neighbors (unless -nsize specified) in each xy plane.
All other voxels set = 0. Max voxels left at initial
intensity (except low order bit is zeroed). This program
uses an "in place" algorithm so no extra storage is needed.
NOTE: a one voxel border is always zeroed out.
Usage: max2d [options] infile.i2i outfile.i2i
options:
-s file: print statistics to "file" of # of maxima found.
-w # : is the width (+/- numpixels) around maxima to include intensity for -s option, default = 1.
-N # : is the width (+/- numpixels) of region examined to determine if a voxel is a max, default = 1.
-t: rather than intensity strictly >, test >= (ie, allow intensity ties).
-T #: only voxels > # in infile.i2i are allowed to be a maxima (i.e, a threshold, default = 0)
-v: verbose
-o orig.i2i: original file to use for output. "infile.i2i"
on the command line may be the original image
cross correlated with a restored bead, and we
are trying to find spots of maximum response.
Once these voxels are identified, however, the
statistics we calculate and the image we manipulate
to create an output image will be "orig.i2i".
-n norm.i2i: normalize the infile.i2i by norm.i2i before
locating maxima. Each voxel in infile.i2i is
divided by the corresponding voxel in norm.i2i
before the location of the maxima is determined.
If infile was the result of blur3d -X to do a
cross correlation (matched filter) then dc
intensity changes in orig.i2i should be compensated
for by the use of this option with norm.i2i an
"averaged" version of orig.i2i.
-f #: factor to multiply the result of the normalization
by before analysis for max (since calculations for
maxima are done on short ints, not floats, the factor is
used to rescale the division back to a reasonable
range (I hope)). Default = 1000.000000
-k: keep a 3 by 3 neighborhood from original image
around each maxima, instead of just the maxima.
Only valid with -o option and without -c option.
-Z: mark voxels which are greater than their neighbors
at least 1 dimension in the xy plane
(so the xy neighbors are checked in 4 subsets:
left-right, up-down, diagonal1, and diagonal2).
-c: output original image, but maxima are set to
-1 (so with play they can show up in color).
NOTE: if -c is used with -z then 3 numbers are
used -4 for zmax, -1 for xymax, and -2 for both.
-r outfile: produce an "rpts" format file for addlines.
note: entire image name is NOT required.
note: a - in place of an image name means stdin or stdout.
see also: max3d
source in /storage/big1/lml/was_vision/colocal
max2d_simple
For each z slice (time point) in the image, find pixels brighter than its 8 neighbors. Print num maxima to stdout.
Only voxels > threshold can be maxima (default = 0.000000). Doesn't use much memory (reads in a slice at a time).
Should handle our regular (short int) image format and floating point images.
If a pixel has no neighbors brighter than it, but some neighbors its same intensity, it will be picked as a maximum
and no other pixels in that plateau will be permitted to be a maxima (thus the pixel chosen as the maximum in
the plateau will depend upon the order pixels are examined).
Usage:
max2d_simple [options] image1.i2i
Options:
-t #: threshold for image1.i2i default = 0.000000
-T #: adaptive threshold. # = pixel intensity/mean intensity for each time pt. must be > # to be maxima.
If -o specified, only pixels under the mask counted calculating mean intensity.
-i thresh.i2i: use with -T option if desired. thresh.i2i is used to calculate the threshold intead of image1.i2i
If -o specified, then that mask will also be applied to thresh.i2i.
-p file.pts: write out coords of maxima to a pts file, w = -1 (use addlines to add back to the image)
-P file.pts #: write out coords of maxima to a pts file, w = # (use addlines to add back to the image)
-p and -P produce extra info on each line after the #, which is comx,comy,comz,iod, and peakval
-q use this with -p or -P to use a quadratic fit instead of comx to get coords and peakval.
-m image.i2i: write out image with all voxels except maxima set to 0. maxima at original intensity.
-M : write out neighborhood around maxima too (must still specify -m). neighborhoods may touch.
-z low high: only analyze z slices from low to high (1-indexed, inclusive). changes zdim of -m image.
-b : pixels along the border are NOT allowed to be maxima
-d #: if (neighbor value + delta) >= pixel value, pixel is NOT a maxima (unless -C). default delta = 0.000000
-e bkgnd num: do NOT allow a pixel to be a maxima if it has >= num neighbors which are
background voxels, that is, whose intensity is <= bkgnd (ie, it is on the edge of a region)
-C #: a pixel can be a max when pixel value > (neighbor value + delta) for at least # neigbhors
(default is 8 neighbors, i.e., all of them).
-n #: neighborhood width. default = 1 (ie, pixel must be >= neighbors within +/- 1 voxel)
-c #: print to stdout, statistics about how the maxima cluster. # is scale factor for dist*sqrt(N) stat.
-o mask.i2i: apply mask image to results, only keep maxima under mask pixels > 0.
mask.i2i must be in our standard image format (short int). If 3d, only first 2d image used.
-v: verbose
See also: pt_distances, max2d, max3dL, maxima, maxL, maxzL, float2intL, int2float, playexact, track
/home/lml/krypton/Satoshi/track_vesicles.m which can read in this format and track the pts.
Example:
maxima must be >100, strictly greater than its neighbors and at most 1 neighbor a bkgnd pixel (<=0)
max2d_simple -t 100 -d .5 -e 0 2 image1.i2i > image1.max
Source code in ~krypton/lml/facil
Copyright 2010 University of Massachusetts Medical School and the Biomedical Imaging Group
All rights reserved. Explicit permission to use this program must be received prior to use.
max3dL
This program finds voxels which are > all 26 neighbors.
All other voxels set = 0. Max voxels left at initial
intensity (except low order bit is zeroed). This program
uses an "in place" algorithm so no extra storage is needed.
NOTE: a one voxel border is always zeroed out.
Usage: max3d [options] infile.i2i outfile.i2i
options:
-s file: print statistics to "file" of # of maxima found.
-t: rather than intensity strictly >, test >= (ie, allow intensity ties).
-o orig.i2i: original file to use for output. "infile.i2i"
on the command line may be the original image
cross correlated with a restored bead, and we
are trying to find spots of maximum response.
Once these voxels are identified, however, the
statistics we calculate and the image we manipulate
to create an output image will be "orig.i2i".
-n norm.i2i: normalize the infile.i2i by norm.i2i before
locating maxima. Each voxel in infile.i2i is
divided by the corresponding voxel in norm.i2i
before the location of the maxima is determined.
If infile was the result of blur3d -X to do a
cross correlation (matched filter) then dc
intensity changes in orig.i2i should be compensated
for by the use of this option with norm.i2i an
"averaged" version of orig.i2i.
-f #: factor to multiply the result of the normalization
by before analysis for max (since calculations for
maxima are done on short ints, not floats, the factor is
used to rescale the division back to a reasonable
range (I hope)). Default = 1000.000000
-k: keep a 3 by 3 neighborhood from original image
around each maxima, instead of just the maxima.
Only valid with -o option and without -c option.
-z: mark voxels which are greater than their neighbors
in z and/or in the xy plane (so only 10 neighbors
are checked in two subsets, 2 z neighbors and 8
neighbors in the xy plane)
-Z: mark voxels which are greater than their neighbors
in z and/or in at least 1 dimension in the xy plane
(so the xy neighbors are checked in 4 subsets:
left-right, up-down, diagonal1, and diagonal2).
-b: only output a max if a voxel is BOTH a max in
z and in xy (possibly only 1d in xy if -Z specified)
This option only needed with -z or -Z options.
If it is not used when -z or -Z is used, then any
voxel which satisfies either max will be output.
-y: mark voxels which are greater than their neighbors
in z.
-c: output original image, but maxima are set to
-1 (so with play they can show up in color).
NOTE: if -c is used with -z then 3 numbers are
used -4 for zmax, -1 for xymax, and -2 for both.
-r outfile: produce an "rpts" format file for addlines.
note: entire image name is NOT required.
note: a - in place of an image name means stdin or stdout.
typical usage:
/*first calculate an "avg" image for normalization later*/
makedcim -x # -y # -z # dcim.i2i /*#'s are the dimensions of psf_r*/
blur3d original dcim avgorig
/*now cross correlate with what you are looking for.*/
blur3d -X original psf_r blurout
/*now normalize the result and find local maxima.*/
max3d -n avgorig.i2i -s bluroutmax.stat -z -c -o original.i2i blurout.i2i bluroutmax.i2i
/*look at results and statistics.*/
play -P -1 R -P -2 G -P -3 B -P -4 Y bluroutmax.i2i
more bluroutmax.stat
maxL
This program finds voxels which are maxima in the z direction
and ALSO maxima in at least one of the four directions in the xy plane
(so the xy neighbors are checked in 4 subsets:
left-right, up-down, diagonal1, and diagonal2).
It is similar to max3d but more restrictive.
All other voxels set = 0. Max voxels left at initial
intensity (except low order bit is zeroed). This program
uses an "in place" algorithm so no extra storage is needed.
NOTE: a one voxel border is always zeroed out.
NOTE: the low order bit of each voxel is used by this program.
Usage: max [options] -o originalfile.i2i infile.i2i outfile.i2i
options:
-s file: print statistics to "file" of # of maxima found.
-o orig.i2i: original file to use for output. "infile.i2i"
on the command line may be the original image
cross correlated with a restored bead, and we
are trying to find spots of maximum response.
Once these voxels are identified, however, the
statistics we calculate and the image we manipulate
to create an output image will be "orig.i2i".
NOTE: this is NECESSARY, it is NOT an option.
NOTE: a voxel which is zero in the original image
is not allowed to be a maximum even if it is
in "infile.i2i".
-n norm.i2i: normalize the infile.i2i by norm.i2i before
locating maxima. Each voxel in infile.i2i is
divided by the corresponding voxel in norm.i2i
before the location of the maxima is determined.
If infile was the result of blur3d -X to do a
cross correlation (matched filter) then dc
intensity changes in orig.i2i should be compensated
for by the use of this option with norm.i2i an
"averaged" version of orig.i2i.
-f #: factor to multiply the result of the normalization
by before analysis for max (since calculations for
are done on short ints, not float, the factor is
used to rescale the division back to a reasonable
range (I hope)). Default = 1000.000000
-k: keep a 3 by 3 neighborhood from original image
around each maxima, instead of just the maxima.
-z: keep zmax only
-r outfile: produce an "rpts" format file for addlines.
note: entire image name is NOT required.
note: a - in place of an image name means stdin or stdout.
typical usage:
/*first calculate an "avg" image for normalization later*/
makedcim -x # -y # -z # dcim.i2i /*#'s are the dimensions of psf_r*/
blur3d original dcim avgorig
/*now cross correlate with what you are looking for.*/
blur3d -X original psf_r blurout
/*now normalize the result and find local maxima.*/
max -s bluroutmax.stat -o original.i2i blurout.i2i bluroutmax.i2i
/*look at statistics.*/
more bluroutmax.stat
You might do this twice, to two separate images and then apply
"overlap" to the two output images to calculate colocalization.
maxzL
This programs find the maximum voxel value in z for each x,y colomn in a 3d data set. The resulting maximum projection onto the xy plane is written out to a 2D image. Usage: maxz [options] input.i2i output.i2i options: none (yet). see also: max, max3d . note: entire image name (with extension) is NOT required. note: a - in place of an image name means stdin or stdout. note: a compressed image will be automatically be uncompressed.
median
Takes a 3d image and median filters each 2d slice. A border of specified width is set to zero. Usage: median [options] inimage.i2i outimage.i2i options: -w #: width of filter region, default = 3. -v: verbose, output to stderr.
planimeter3
/home/lml/krypton/bin/planimeter3: /usr/lib/libGL.so.1: no version information available (required by /home/lml/krypton/bin/planimeter3)
PLANIMETER3: Measure length and area of closed contours
Usage:
planimeter3 [options]
Options:
-I input_file Image file name(s). A maximum of three
images can be entered. The first one is displayed
as green, the second as red and the third as blue.
Voxels colocalised between any two images are shown
as white
-R rpts file. If it exists on input, it is read in and
contours contained in it are displayed and can be deleted
or edited. If it does not exist, it will be saved on
exit unless Quit is pressed first
-S n n n Image number, display scale and black-level
-X n n range of X planes (low,high)
-Y n n range of Y planes (low,high)
-Z n n range of Z planes (low,high)
-M n Magnification (zoom)
This program allows you to draw boundaries around a cell or object
The boundaries can be drawn in any order and can be erased if required.
Use:
Left mouse button: move between planes (as in play).
Middle mouse button: draw boundary.
Once a object has been defined pressing the middle mouse
button will start drawing a contour for that object, or,
if the contour has already been started, continue it.
Each subsequent perimeter is associated with the previously
defined object. To change the object or start a new object
use the pop-up menu.
Backspace - undraw previous point (may be repeated).
Keypad + : Zoom in.
Keypad - : Zoom out.
l - (toggle) show n planes in addition to the current plane.
Where n is 3 (default) but can be set from 1 - 9 by pressing
the appropriate numerical key.
Pressing the l key again will show just the current plane
v - (toggle) while drawing will result in a vertical line being
drawn from the current position when the mouse is moved.
h - (toggle) while drawing will result in a horizontal line being
drawn from the current position when the mouse is moved.
Control (toggle) - Toggle on and off scaling for the images.
Notes: A maximum of five separate objects can be defined for
contouring. Each object is represented by a different
colour.
The only limit on the number of contours that may
added to an object is the number of Z planes.
There is a limit of 50000 points per drawn contour,
however there is no limit on the number of points
of a contour in a file that is read-in
play_huge
This program displays huge short int images. It only reads in a "window" of images at a time. If you display past the range of images stored in the window, the next z plane is automatically read and the oldest (smallest z) plane automatically dropped. Right now you are not allowed to back up past the oldest plane in the window. It can also print out pixel values in a small region surrounding the cursor. These may be approximate values. note: modified from playexact Usage: play_huge [options] image.i2i options: -S #1 #2: rescale numbers for display (0-255) as new = (old - #2)/#1, ie, #1 is the scale factor and #2 the background. default is to use the initial window of images to autoscale. -s #: start reading images at z plane # (1-indexed) -w #: the number of zplanes in the window of planes read in, default = 1 -f path size the full path (including file name) of the font to use and pointsize of font default: /usr/X11R6/lib/X11/fonts/Type1/times.ttf 12. /usr/share/fonts is another common location. -x namedfifo: read location information from the named fifo periodically. If the namedfifo doesn't exist it will be created (and deleted upon exit from the "Quit" menu entry). Alternatively it can be created ahead of time using mkfifo. The exo program produces output to stdout. found.pl -trackxmgr converts this to an xmgr file. xmgrace can display this file. xmgrace->window->command: WRITE G0.s0 FILE "namedfifo" will send the info to the namedfifo. play_huge will then see this and jump to that location. Currently only works with modified krypton:/usr/local/grace/bin/xmgrace program. -g grace_path : by default "xmgrace" is called. On krypton this is /usr/local/grace/bin/xmgrace, which has the mods to handle the -x option, but it is buggy. A better version is /usr/X11R6/bin/xmgrace. -v: verbose -V: more verbose note: a - in place of an image name means stdin or stdout. Once in the program, move forward or back in z by pressing the f or b key, respectively. Help = h key. H: switch output values to hex format. D: switch output values back to decimal format. Other capabilities may also be present, see h key info. Source in /home/lml/krypton/facil See Also: Examples:
printvalsL
#/home/lml/krypton/bin/printvalsL
Prints (x,y,z,count,value) to stdout. Count is
how many values have been printed (see -c option). Value is the image value.
Coords are 1-indexed. Prints values over a range of coords. If a coordinate is not
specified then all values along that dimension are used.
This version handles both short integer and floating point (not extensively tested) images.
Usage: printvalsL [options] image.i2i
options:
-x # #: x-coords to print (low high), 1-indexed, default is all x coords.
-y # #: y-coords to print, 1-indexed, default is all y coords.
-z # # #: z-coords to print (low hi delta), 1-indexed, default is all z coords.
-m: print the middle (x,y) value, all z values.
-e: print the values of brightest pixel in volume (all pixels at same x y position).
-E: print the values of biggest dF/F pixel in volume (all pixels at same x y position).
(note, still need -d if the value you want to print is the dF/F value, -E just picks
the location to print based upon dF/F)
-X range: modifies -m,-e, or -E, takes the position found and +- range (in pixels) in x.
-Y range: same but for y coord.
-d: print dFoverF, ie, 100*scale*(val-val0)/val0 where val0 is the first
value printed. Not correct if z = 0 excluded (-z option) or multiple xy points printed.
-s scale: set scale in the above equation, default = 1.000000
-rx range: set a range of x coords to average over (eg. 2 means +- 2, a 5 pixel region).
-ry range: same for y.
-rz range: same for z.
-R: radial coords as in nadiffl7. Scale voxels based upon volume in this coord system.
(so value -> 2pi*r*dr*dl*value). By default this only applies to the weights when averaging
a range of values (-rx,-ry, or -rz options), if you want it to apply to regular values, you
must specify the -t option also.
-S: x coord is scaled as if in sperical coords (ie, value -> 4pi*r*r*dr*dl*value). also use -t option
if you want this to apply to regular values too.
-p filename: read pixel size (spacing file) from filename (see cyl2cart2 also, and cellsim).
-t: apply the -R or -S option to individual pixel values, not just when weighting to find a local average.
-c start scale: start number (integer, default = 1) and scale for first number of pair.
-i: print out slice, IOD, IOD/volume for each zslice and total iod for the image.
Can be combined with -R, -d.
-x, -y, -z, modifies this to just calculate iod for a subset of the image
(as does -m|-e|-E and -X or -Y).
-b #: ignore any value less than base #, default is to accept all values
(nadiffl7 sometimes creates negative numbers via overflow)
-o file: name of file (eg, fluo3.dat) which contains true values of pixels which had
overflow. This is for output from nadiffl7 (which writes 32767 at overflow pixels).
The true values will be read from this file.
-O x y z: x y and z offsets to add to coords to use -o option. This is only needed if the image
passed to printvals is not the same size as when the overflow file (eg,fluo3.dat) was
produced. So if the y = 1 was segmented off and the first 3 z slices use: -O 0 1 3
Note:if you segmented off the high end, ignore that, since it doesn't affect remaining coords.
note: entire image name (with extension) is NOT required.
note: a - in place of an image name means stdin or stdout.
note: a compressed image will be automatically be uncompressed.
examples:
#want to take 100nm simulation and produce dFoverF values as if we had 500 nm voxels
#only want the central (x,y) coord since that is where the calcium enters the cell
#the simulation (ie, each z zslice) was every 10 msec, but we want output in msec
printvalsL -m -rx 2 -ry 2 -d -s 100 -c 0 10 file.i2i
#want to know how fast the total free stuff in the cell changes (simulation started at t=4200msec
#and output to cylindrical coords was every msec)
printvalsL -i -d -c 4200 1 -R file.i2i
#want to calculate the total change in light (a proxy for delta_calcium) in a 21x21 region
printvalsL -i -e -X 10 -Y 10 file.i2i
[source in krypton:/home/lml/krypton/facil/printvals.cc]
running_avg
Lets you performing a temporal (i.e., in z) running average. It only reads a few z slices at a time so doesn't use much memory. Reads and writes sequentially without closing a file, so hopefully should work on pipes too. This smooths data, and also helps eliminate noise. Usage: running_avg [options] inimage.i2i outimage.i2i Options: -r minx miny maxx maxy: only average within this region (inclusive, 1-indexed). -z startz stopz zw: for each z slice between z = startz and z = stopz (inclusive, 1-indexed) calculates: avg(z-zw, ..., z+zw). if startz = -1, that means zw+1. if stopz = -1, that means zdim-zw. default = -z -1 -1 1 -t: keep the zw slices from the beginning and end (so outimage.i2i same zdim as inimage.i2i) So if zw = 2, slices from 3 to end-2 (1-indexed) will be an average of 5 slices (as usual) But slice 1 will be the avg of 3 slices (1...3), and slice 2 the average of 4 slices (z=1...4) -d: extra debugging info -v: verbose Examples: running_avg -z -1 -1 3 inimage.i2i outimage.i2i : +/-3 zplane running average Bugs: the created image has a strange permission. fix it by doing: chmod 644 outimage.i2i Source code in ~krypton/lml/facil
running_mult
Lets you perform a temporal (i.e., in z) running multiply. It only reads a few (zw) z slices at a time so doesn't use much memory. Reads and writes sequentially without closing a file, so hopefully should work on pipes too. Use this to see which pixels in a mask persist over a period of time. By default converts image to binary (pixels > threshold set = 1) and produces a binary image. This routine might be used to only keep objects (pixels) which have persisted (been above threshold) for at least a certain amount of time (i.e., zw zslices worth, where z is time). For example, finding which clathrin vesicles (for Silvia) are stationary for at least 1 minute. Usage: running_mult [options] inimage.i2i outimage.i2i Options: -r minx miny maxx maxy: only average within this region (inclusive, 1-indexed). -z startz stopz zw: for each z slice between z = startz and z = stopz (inclusive, 1-indexed) calculates: mult(z-zw +1, ... z), i.e, multiplies zw slices preceding (and including) z. if startz = -1, that means zw+1. if stopz = -1, that means zdim. default = -z -1 -1 1 -k: keep the slices before zstart and after zstop (so outimage.i2i same zdim as inimage.i2i) -t #: pixels > # are above threshold (set to 1), default = 0. -o: instead of writing out a binary image, apply binary mask to original image values. -m: median filter instead of multiply. This is more robust than multiplication, so if an object moves a bit and then moves back quickly, it will be preserved. Slow. only works with our standard image format (short int). Pixels <= threshold considered 0. -e #: max filter instead of multiply. This works like reduceima -Z start stop zw I think. # is the value to which pixels outside the region (if -r option) or <= threshold are set. -d: extra debugging info -v: verbose Examples: running_mult -z -1 -1 3 inimage.i2i outimage.i2i : 3 zplane running multiply Bugs: the created image has a strange permission. fix it by doing: chmod 644 outimage.i2i Source code in ~krypton/lml/facil
scratchng_bg
For each z slice in the image subtract off the dark current pixel value, write out a new image. The dark current for each of 8 quadrants in the image (i.e., 8 ports, 2 rows by 4 columns) is the average of all the dark current pixels for that quadrant (the last 8 columns of pixels are assumed to be the dark current values, 2 pixels wide for each port). The new image will have an xdim 8 less than the original (since the dark current pixels are removed). Pixel values are allowed to go negative. Doesn't use much memory (since it reads in a z slice at a time). Should handle our regular (short int) image format and floating point images. May not correctly set tdim in resulting image Usage: scratchng_bg [options] original.i2i new.i2i Options: -v: verbose See also: /storage/big1/kef/Linux/bin/scratchng - should be the same, but doesn't read in a zslice at a time Examples: Source code in ~krypton/lml/facil
temporal_projection
Lets you project pixels in z, producing a single 2d image. Any pixel greater than threshold (see -t) will be projected. Each projected pixel just has its z value (1-indexed) projected, NOT its intensity. Thus the input image can be thought of as a binary image, and the projected image has brighter pixels represent pixels later in z (time). Reads one z plane at a time, so should work on large images, also floating point images. Usage: temporal_projection [options] inimage.i2i outimage.i2i Options: -z startz stopz: only project within the range of z slices (inclusive, 1-indexed). default = all z slices. -t #: pixels > # are above threshold and therefore projected, default = 0. -o: instead of keeping the newest (largest z) pixel, keep the oldest. -b: instead of keeping the newest (largest z) pixel, keep the brightest. -m #: for -b, set initial background values to # instead of 0 -p: istead of writing out the zvalue, write out the pixel value -s file.i2i: for each projected slice, save the built up image, so file.i2i will have stopz-startz+1 zdim -d: extra debugging info -v: verbose Examples: temporal_projection inimage.i2i outimage.i2i Source code in ~krypton/lml/facil
zeros
<nowiki>
This program finds the zero crossings of an image. Pixels at the zero crossing are set to 1. Others are zero. 3D images have each z slice examined separately. It can read in REAL or SHORT INT images. It writes out a SHORT INT image (numbers too big or small are set to MAX or MIN).
Usage: zeros [options] inimage.i2i outimage.i2i options: -c: ensure that the contour is closed (unless it touches the image border). This causes thick contours but is useful for later processing (e.g., by a flood fill). -z #: set the value of a pixel at a zero crossing to be # (instead of 1) -p: set the value of a zero crossing pixel to be proportional (equal) to positive pixel minus the negative pixel value at the zero crossing. NOTE: any output pixel value <1 for a zero crossing will be set to 1. note: entire image name (with extension) is NOT required. note: a - in place of an image name means stdin or stdout.
<nowiki>