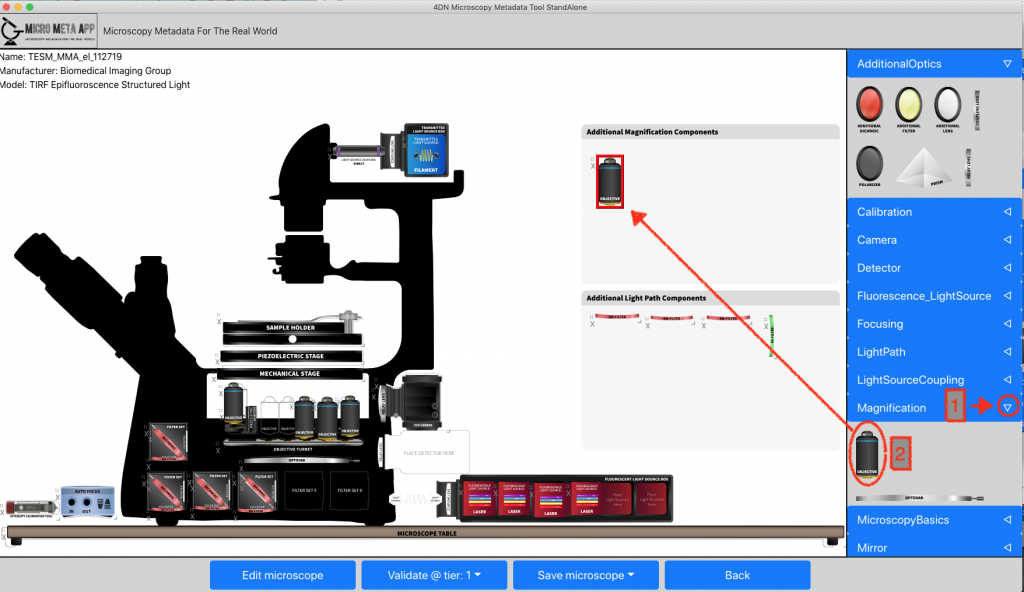
Micro-Meta App is designed to aid in the collection of both Microscope Hardware Specifications and Image Acquisition Settings metadata. In this example, a previously saved Microscope file was selected from an available repository and opened for further editing. In order to add the metadata associated with a newly purchased objective to a Microscope file the “Magnification” drop-down menu is opened [1] and an additional “Objective” [2] is dragged onto the workspace.
Micro-Meta App facilitates the collection of both Microscope Hardware Specifications and Image Acquisition Settings metadata on the basis of the proposed 4DN extension of the OME Core model, which was recently posted as a white paper on ArXiv.org. A beta version of Micro-Meta App was released in December 2019.
Available implementations of Micro-Meta App include:
- A web app version implemented in Javascript React
- A stand-alone version implemented in Javascript Electron
- A prototype plugin version to be integrated into the OMERO.web browser
Background:
For the information content of image data to be unequivocally extracted and machine-readable, microscopy images need to be accompanied by thorough documentation of the microscope hardware, imaging settings, and instrument performance that ensure the correct interpretation of results. A significant challenge with the reproducibility of microscopy results and with their integration with chromatin folding maps generated by the 4DN Consortium lies in the lack of universally accepted super-resolution microscopy quality control and reporting standards and of widely available cyberinfrastructure to support the collection of provenance metadata. To address this challenge, the 4DN consortium has put forth a 4DN extension of the OME Core metadata model, which includes a tiered system of reporting guidelines that scales quality control and reporting requirements with experimental complexity.
Summary:
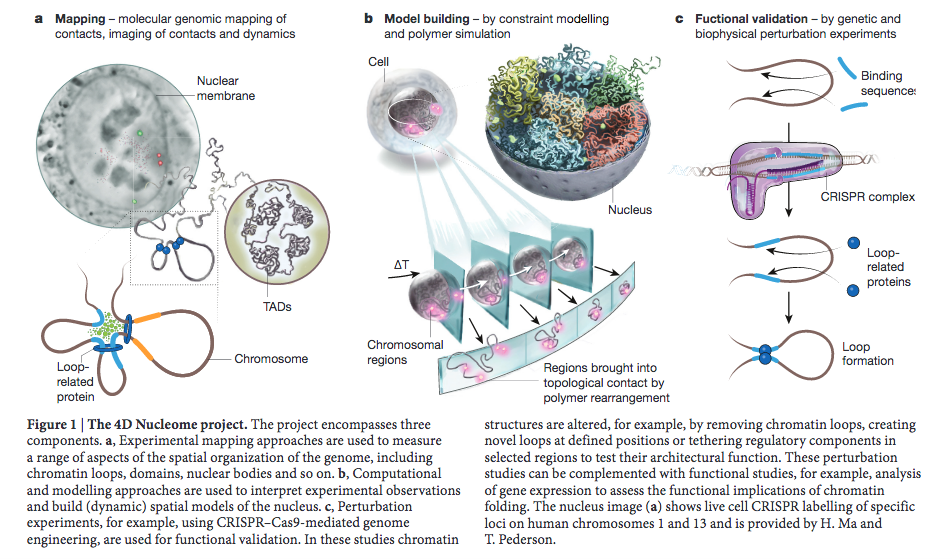
Micro-Meta App is a novel application that provides an interactive and intuitive approach for rigorous record-keeping in fluorescence microscopy and is based on the 4DN-OME Microscopy metadata standard and on the proposed tiered-system of guidelines. The user’s data processing workflow consists of multiple steps. First, in the Create Microscope modality (Figure 1), the App allows the users to build a graphical representation of the microscope hardware by dragging-and-dropping individual components onto the workspace and entering the relevant attribute values based on the selected tier level. Second, Micro-Meta App generates tier-specific descriptions of the microscope hardware and exports them in a Microscope file that can be used as a template and shared with the community, with a significant reduction in the recordkeeping burden. Then, in the Use Microscope modality, Micro-Meta App opens an existing Microscope file, imports Image Acquisition settings from the header of image data files to be annotated and interactively guides the user through the collection of all missing instrument-specific and tier-appropriate image acquisition settings and calibration metrics required to ensure reuse and reproducibility of image data. Finally, the App generates JSON files that contain comprehensive descriptions of the conditions utilized to produce individual microscopy datasets, and that can be stored on the user’s file system, or on third party repositories. To lower the barrier of adoption of Micro-Meta App by a wide community of users the application is available as a stand-alone program, as a plugin of the OMERO web client and as a service of the 4DN data portal.